8EYE
 
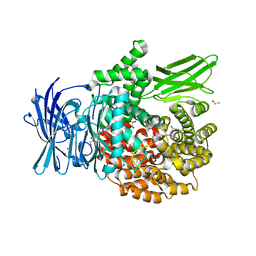 | | Plasmodium falciparum M1 in complex with inhibitor 9aj | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
7PIH
 
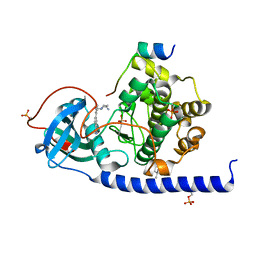 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN093 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[3-[[(3~{R})-3-azanylpyrrolidin-1-yl]methyl]phenyl]-4~{H}-isoquinolin-1-one, CHLORIDE ION, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
8EWZ
 
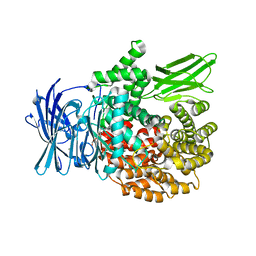 | | Plasmodium falciparum M1 in complex with inhibitor 9c | | Descriptor: | (2R)-2-(cyclopentylcarbamamido)-N-hydroxy-2-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)acetamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
7PNS
 
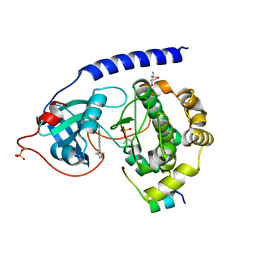 | | Protein kinase A catalytic subunit in complex with PKI5-24 and EN081 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-azanyl-6-[5-[(dimethylamino)methyl]-2-fluoranyl-phenyl]-1H-indole-3-carbonitrile, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Glinca, S, Mueller, J.M, Ruf, M, Merkl, S. | | Deposit date: | 2021-09-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.855 Å) | | Cite: | Magnet for the Needle in Haystack: "Crystal Structure First" Fragment Hits Unlock Active Chemical Matter Using Targeted Exploration of Vast Chemical Spaces.
J.Med.Chem., 65, 2022
|
|
8EKH
 
 | | I-2 Y35N H35N (unbound) Fab from CH65-CH67 lineage | | Descriptor: | I-2 Fab heavy chain, I-2 Fab light chain | | Authors: | Maurer, D.P, Schmidt, A.G. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hierarchical sequence-affinity landscapes shape the evolution of breadth in an anti-influenza receptor binding site antibody.
Elife, 12, 2023
|
|
8EZ2
 
 | | Plasmodium falciparum M1 in complex with inhibitor 15ag | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
8EYF
 
 | | Plasmodium falciparum M1 in complex with inhibitor 15aa | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
6D04
 
 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; two molecules of parasite ligand, subclass 1. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
8EXG
 
 | | Human Carbonic Anhydrase II bound N-(2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethyl)-4-sulfamoylbenzamide | | Descriptor: | (4S)-5-amino-4-{1,3-dioxo-4-[(2-{2-[2-(4-sulfamoylbenzamido)ethoxy]ethoxy}ethyl)amino]-1,3-dihydro-2H-isoindol-2-yl}-5-oxopentanoic acid, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
8EXC
 
 | | Human Carbonic Anhydrase II bound tert-butyl (3-(4-(3-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)propoxy)butoxy)propyl)carbamate | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, N-(3-{4-[3-({2-[(3R)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}amino)propoxy]butoxy}propyl)-4-sulfamoylbenzamide, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
8EJB
 
 | |
8EMU
 
 | |
8EYL
 
 | | Human Carbonic Anhydrase II with Tert-butyl (2-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethyl)carbamate | | Descriptor: | 2-[[(2~{R})-1-azanyl-5-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]carbamoyl]-6-[2-[2-[(4-sulfamoylphenyl)carbonylamino]ethoxy]ethylamino]benzoic acid, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Kohlbrand, A.J, O'Herin, C.B. | | Deposit date: | 2022-10-27 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Development of Human Carbonic Anhydrase II Heterobifunctional Degraders.
J.Med.Chem., 2023
|
|
8TH9
 
 | |
8T21
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein | | Descriptor: | Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T22
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors at downRBD conformation | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8TAZ
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to one mink ACE2 receptors | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Liang, B. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T20
 
 | | Cryo-EM structure of mink variant Y453F trimeric spike protein bound to two mink ACE2 receptors | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T25
 
 | | Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at downRBD conformation. | | Descriptor: | Angiotensin-converting enzyme, Spike glycoprotein | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8T23
 
 | | Cryo-EM structure of the RBD-ACE2 interface of the SARS-CoV-2 trimeric spike protein bound to ACE2 receptor after local refinement at upRBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Ahn, H.M, Calderon, B, Fan, X, Gao, Y, Horgan, N, Zhou, B, Liang, B. | | Deposit date: | 2023-06-05 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural basis of the American mink ACE2 binding by Y453F trimeric spike glycoproteins of SARS-CoV-2.
J Med Virol, 95, 2023
|
|
1TVU
 
 | |
6Z1M
 
 | | Structure of an Ancestral glycosidase (family 1) bound to heme | | Descriptor: | 1,2-ETHANEDIOL, Ancestral reconstructed glycosidase, GLYCEROL, ... | | Authors: | Gavira, J.A, Risso, V.A, Sanchez-Ruiz, J.M, Gamiz-Arco, G, Gutierrez-Rus, L, Ibarra-Molero, B, Oshino, Y, Petrovic, D, Romero-Rivera, A, Seelig, B, Kamerlin, S.C.L, Gaucher, E.A. | | Deposit date: | 2020-05-14 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Heme-binding enables allosteric modulation in an ancient TIM-barrel glycosidase.
Nat Commun, 12, 2021
|
|
7Q5S
 
 | | Protein community member fatty acid synthase complex from C. thermophilum | | Descriptor: | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase, 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Chojnowski, G, Skalidis, I, Kyrilis, F.L, Tueting, C, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2021-11-04 | | Release date: | 2022-02-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Cryo-EM and artificial intelligence visualize endogenous protein community members.
Structure, 30, 2022
|
|
2THF
 
 | | STRUCTURE OF HUMAN ALPHA-THROMBIN Y225F MUTANT BOUND TO D-PHE-PRO-ARG-CHLOROMETHYLKETONE | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, SODIUM ION, THROMBIN HEAVY CHAIN, ... | | Authors: | Caccia, S, Futterer, K, Di Cera, E, Waksman, G. | | Deposit date: | 1999-01-26 | | Release date: | 1999-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected crucial role of residue 225 in serine proteases.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8SYJ
 
 | | Structure of apurinic/apyrimidinic DNA lyase TK0353 from Thermococcus kodakarensis (Iodide crystal form) | | Descriptor: | GLYCEROL, IODIDE ION, SULFATE ION, ... | | Authors: | Eckenroth, B.E, Gardner, A.F, Doublie, S. | | Deposit date: | 2023-05-25 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thermococcus kodakarensis TK0353 is a novel AP lyase with a new fold.
J.Biol.Chem., 300, 2023
|
|
