6TPR
 
 | | PqsR (MvfR) bound to inhibitory compound 40 | | Descriptor: | 2-[(5-methyl-[1,2,4]triazino[5,6-b]indol-3-yl)sulfanyl]-~{N}-(4-pyridin-2-yloxyphenyl)ethanamide, Transcriptional regulator MvfR | | Authors: | Richardson, W.K, Emsley, J. | | Deposit date: | 2019-12-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hit Identification of New Potent PqsR Antagonists as Inhibitors of Quorum Sensing in Planktonic and Biofilm GrownPseudomonas aeruginosa.
Front Chem, 8, 2020
|
|
4BQH
 
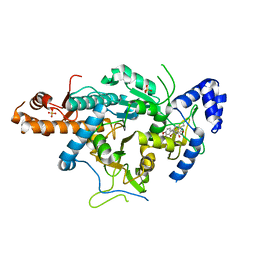 | | Crystal structure of the uridine diphosphate N-acetylglucosamine pyrophosphorylase from Trypanosoma brucei in complex with inhibitor | | Descriptor: | (3S)-3-[2-(1,3-benzodioxol-5-yl)-2-oxidanylidene-ethyl]-4-bromanyl-5-methyl-3-oxidanyl-1H-indol-2-one, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Fang, W, Raimi, O.G, vanAalten, D.M.F. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Novel Allosteric Inhibitor of the Uridine Diphosphate N-Acetylglucosamine Pyrophosphorylase from Trypanosoma Brucei.
Acs Chem.Biol., 8, 2013
|
|
6TQY
 
 | |
1EBC
 
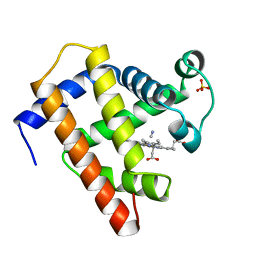 | | SPERM WHALE MET-MYOGLOBIN:CYANIDE COMPLEX | | Descriptor: | CYANIDE ION, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rosano, C, Ascenzi, P, Rizzi, M, Losso, R, Bolognesi, M. | | Deposit date: | 1999-03-04 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyanide binding to Lucina pectinata hemoglobin I and to sperm whale myoglobin: an x-ray crystallographic study.
Biophys.J., 77, 1999
|
|
1G7G
 
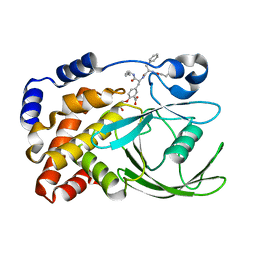 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXES WITH PNU179326 | | Descriptor: | 2-(CARBOXYMETHOXY)-5-[(2S)-2-({(2S)-2-[(3-CARBOXYPROPANOYL)AMINO] -3-PHENYLPROPANOYL}AMINO)-3-OXO-3-(PENTYLAMINO)PROPYL]BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Bleasdale, J.E, Ogg, D, Larsen, S.D. | | Deposit date: | 2000-11-10 | | Release date: | 2001-06-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small molecule peptidomimetics containing a novel phosphotyrosine bioisostere inhibit protein tyrosine phosphatase 1B and augment insulin action.
Biochemistry, 40, 2001
|
|
6TM0
 
 | | N-Domain P40/P90 Mycoplasma pneumoniae complexed with 6'SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Vizarraga, D, Aparicio, D, Illanes, R, Fita, I, Perez-Luque, R, Martin, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae.
Nat Commun, 11, 2020
|
|
4D43
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 2-(2- chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(2-chloro-4-nitrophenoxy)-5-ethyl-4-fluorophenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
6TNR
 
 | | PI3K delta in complex with N[5(7{2[4(2hydroxypropan2yl)piperidin1 yl]ethoxy}1,3dihydro2benzofuran5yl)2 methoxypyridin3yl]methanesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[2-methoxy-5-[7-[2-[4-(2-oxidanylpropan-2-yl)piperidin-1-yl]ethoxy]-1,3-dihydro-2-benzofuran-5-yl]pyridin-3-yl]methanesulfonamide | | Authors: | Convery, M.A, Rowland, P, Henley, Z.A, Barton, N, Down, K. | | Deposit date: | 2019-12-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of Orally Bioavailable PI3K delta Inhibitors and Identification of Vps34 as a Key Selectivity Target.
J.Med.Chem., 63, 2020
|
|
4D44
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-ethyl- 4-fluoro-2-((2-fluoropyridin-3-yl)oxy)phenol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 5-ethyl-4-fluoro-2-[(2-fluoropyridin-3-yl)oxy]phenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2014-10-26 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Ordered Water Channel in Staphylococcus Aureus Fabi: Unraveling the Mechanism of Substrate Recognition and Reduction.
Biochemistry, 54, 2015
|
|
6TQW
 
 | |
1FTP
 
 | |
6TS6
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[[3-[(3~{S})-3-azanyl-2,3-dihydro-1-benzofuran-5-yl]-5-(2-cyanopropan-2-yl)phenyl]methoxy]phenyl]ethanoic acid, Coagulation factor XI, DIMETHYL SULFOXIDE, ... | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
4CSO
 
 | | The structure of OrfY from Thermoproteus tenax | | Descriptor: | ORFY PROTEIN, TRANSCRIPTION FACTOR | | Authors: | Zeth, K, Hagemann, A, Siebers, B, Martin, J, Lupas, A.N. | | Deposit date: | 2014-03-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
4AXF
 
 | | InsP5 2-K in complex with Ins(3,4,5,6)P4 plus AMPPNP | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, Myo inositol 3,4,5,6 tetrakisphosphate, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
4GKX
 
 | | Crystal structure of a carbohydrate-binding domain | | Descriptor: | CALCIUM ION, GLYCEROL, Protein ERGIC-53, ... | | Authors: | Page, R.C, Zheng, C, Nix, J.C, Misra, S, Zhang, B. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Characterization of Carbohydrate Binding by LMAN1 Protein Provides New Insight into the Endoplasmic Reticulum Export of Factors V (FV) and VIII (FVIII).
J.Biol.Chem., 288, 2013
|
|
4B7X
 
 | | Crystal structure of hypothetical protein PA1648 from Pseudomonas aeruginosa. | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROBABLE OXIDOREDUCTASE | | Authors: | Alphey, M.S, McMahon, S.A, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4AVY
 
 | | The AEROPATH project and Pseudomonas aeruginosa high-throughput crystallographic studies for assessment of potential targets in early stage drug discovery. | | Descriptor: | PROBABLE SHORT-CHAIN DEHYDROGENASE | | Authors: | Moynie, L, McMahon, S.A, Alphey, M.S, Liu, H, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-05-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4B79
 
 | | THE AEROPATH PROJECT AND PSEUDOMONAS AERUGINOSA HIGH-THROUGHPUT CRYSTALLOGRAPHIC STUDIES FOR ASSESSMENT OF POTENTIAL TARGETS IN EARLY STAGE DRUG DISCOVERY. | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROBABLE SHORT-CHAIN DEHYDROGENASE | | Authors: | Moynie, L, McMahon, S.A, Alphey, M.S, Liu, H, Duthie, F, Naismith, J.H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1G7F
 
 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH PNU177496 | | Descriptor: | 2-{4-[(2S)-2-[({[(1S)-1-CARBOXY-2-PHENYLETHYL]AMINO}CARBONYL)AMINO]-3-OXO-3-(PENTYLAMINO)PROPYL]PHENOXY}MALONIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Bleasdale, J.E, Ogg, D, Larsen, S.D. | | Deposit date: | 2000-11-10 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small molecule peptidomimetics containing a novel phosphotyrosine bioisostere inhibit protein tyrosine phosphatase 1B and augment insulin action.
Biochemistry, 40, 2001
|
|
7LSW
 
 | | Structure of Full Beta-Hairpin LIR from FNIP2 Bound to GABARAP | | Descriptor: | Folliculin-interacting protein 2,Gamma-aminobutyric acid receptor-associated protein, SULFATE ION | | Authors: | Appleton, B.A. | | Deposit date: | 2021-02-18 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | GABARAP sequesters the FLCN-FNIP tumor suppressor complex to couple autophagy with lysosomal biogenesis.
Sci Adv, 7, 2021
|
|
4DUS
 
 | | Structure of Bace-1 (Beta-Secretase) in complex with N-((2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-((6'-neopentyl-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl)amino)butan-2-yl)acetamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Potent and Orally Efficacious, Hydroxyethylamine-Based Inhibitor of beta-Secretase.
ACS Med Chem Lett, 3, 2012
|
|
4DMK
 
 | |
4DDO
 
 | |
1E9N
 
 | | A second divalent metal ion in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1, and its implications for the catalytic mechanism | | Descriptor: | DNA-(APURINIC OR APYRIMIDINIC SITE) LYASE, LEAD (II) ION | | Authors: | Beernink, P.T, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-10-24 | | Release date: | 2001-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism
J.Mol.Biol., 307, 2001
|
|
1EV1
 
 | | ECHOVIRUS 1 | | Descriptor: | ECHOVIRUS 1, MYRISTIC ACID, PALMITIC ACID | | Authors: | Wien, M.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-12-02 | | Release date: | 1999-01-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure determination of echovirus 1.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
