1TZ6
 
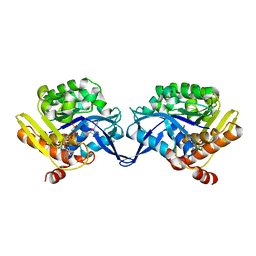 | | Crystal structure of aminoimidazole riboside kinase from Salmonella enterica complexed with aminoimidazole riboside and ATP analog | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOSIDE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Dougherty, M, Downs, D.M, Ealick, S.E. | | Deposit date: | 2004-07-09 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of an Aminoimidazole Riboside Kinase from Salmonella enterica; Implications for the Evolution of the Ribokinase Superfamily
STRUCTURE, 12, 2004
|
|
3SD2
 
 | |
8B27
 
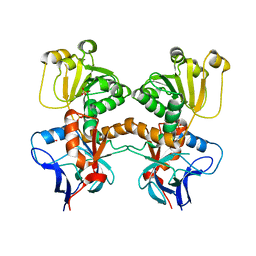 | | Dihydroprecondylocarpine acetate synthase from Catharanthus roseus | | Descriptor: | Dehydroprecondylocarpine acetate synthase, SULFATE ION | | Authors: | Langley, C, Basquin, J, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-09-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1TZJ
 
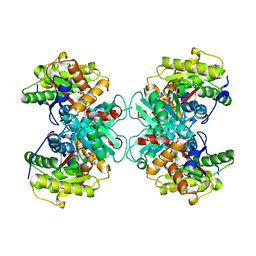 | | Crystal Structure of 1-aminocyclopropane-1-carboxylate deaminase complexed with d-vinyl glycine | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, D-VINYLGLYCINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-10 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
8B8J
 
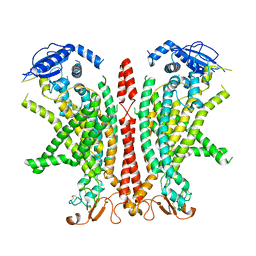 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
1U0E
 
 | | Crystal structure of mouse phosphoglucose isomerase | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Glucose-6-phosphate isomerase, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
3SDI
 
 | | Structure of yeast 20S open-gate proteasome with Compound 20 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N~4~-(2,2-dimethylpropyl)-N~1~-{(2S)-1-[(4-methylbenzyl)amino]-1-oxo-4-phenylbutan-2-yl}-N~2~-[(5-methyl-1,2-oxazol-3-yl)carbonyl]-L-aspartamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2011-06-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8AXC
 
 | | Crystal structure of mouse Ces2c | | Descriptor: | Acylcarnitine hydrolase, CHLORIDE ION, NICOTINAMIDE, ... | | Authors: | Eisner, H, Riegler-Berket, L, Rodriguez Gamez, C, Sagmeister, T, Chalhoub, G, Darnhofer, B, Panikkaveetil Jawaharlal, J, Birner-Gruenberger, R, Pavkov-Keller, T, Haemmerle, G, Schoiswohl, G, Oberer, M. | | Deposit date: | 2022-08-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Crystal Structure of Mouse Ces2c, a Potential Ortholog of Human CES2, Shows Structural Similarities in Substrate Regulation and Product Release to Human CES1.
Int J Mol Sci, 23, 2022
|
|
1UFJ
 
 | | Crystal Structure of an Artificial Metalloprotein:Fe(III)(3,3'-Me2-salophen)/apo-A71G Myoglobin | | Descriptor: | 'N,N'-BIS-(2-HYDROXY-3-METHYL-BENZYLIDENE)-BENZENE-1,2-DIAMINE', FE (III) ION, MYOGLOBIN, ... | | Authors: | Ueno, T, Ohashi, M, Kono, M, Kondo, K, Suzuki, A, Yamane, T, Watanabe, Y. | | Deposit date: | 2003-05-30 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Artificial Metalloproteins: Tight Binding of Fe(III)(Schiff-Base) by Mutation of Ala71 to Gly in Apo-Myoglobin
Inorg.Chem., 43, 2004
|
|
2IHY
 
 | | Structure of the Staphylococcus aureus putative ATPase subunit of an ATP-binding cassette (ABC) transporter | | Descriptor: | ABC transporter, ATP-binding protein, SULFATE ION | | Authors: | McGrath, T.E, Yu, C.S, Romanov, V, Lam, R, Dharamsi, A, Virag, C, Mansoury, K, Thambipillai, D, Richards, D, Guthrie, J, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2006-09-27 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Staphylococcus aureus putative ATPase subunit of an ATP-binding cassette (ABC) transporter
To be Published
|
|
8BC0
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518A Q623A mutant in GDN open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
1U0V
 
 | | An Aldol Switch Discovered in Stilbene Synthases Mediates Cyclization of Specificity of Type III Polyketide Synthases: 18xCHS structure | | Descriptor: | Chalcone synthase 2 | | Authors: | Austin, M.B, Bowman, M.E, Ferrer, J.-L, Schroder, J, Noel, J.P. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Aldol Switch Discovered in Stilbene Synthases Mediates Cyclization Specificity of Type III Polyketide Synthases
Chem.Biol., 11, 2004
|
|
2ID0
 
 | | Escherichia coli RNase II | | Descriptor: | Exoribonuclease 2, MANGANESE (II) ION | | Authors: | Zuo, Y, Zhang, J, Wang, Y, Malhotra, A. | | Deposit date: | 2006-09-13 | | Release date: | 2006-10-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for Processivity and Single-Strand Specificity of RNase II.
Mol.Cell, 24, 2006
|
|
3SEK
 
 | | Crystal Structure of the Myostatin:Follistatin-like 3 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Follistatin-related protein 3, Growth/differentiation factor 8 | | Authors: | Cash, J.N, Thompson, T.B. | | Deposit date: | 2011-06-10 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structure of myostatinfollistatin-like 3: N-terminal domains of follistatin-type molecules exhibit alternate modes of binding.
J.Biol.Chem., 287, 2012
|
|
8B2X
 
 | |
1U1E
 
 | | Structure of e. coli uridine phosphorylase complexed to 5(phenylseleno)acyclouridine (PSAU) | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-(PHENYLSELANYL)PYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Bu, W, Settembre, E.C, Ealick, S.E. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U21
 
 | | transthyretin with tethered inhibitor on one monomer. | | Descriptor: | 2-[(3,5-DICHLORO-4-TRIOXIDANYLPHENYL)AMINO]BENZOIC ACID, Transthyretin | | Authors: | Wiseman, R.L, Johnson, S.M, Kelker, M.S, Foss, T, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2004-07-16 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Kinetic stabilization of an oligomeric protein by a single ligand binding event
J.Am.Chem.Soc., 127, 2005
|
|
1UFQ
 
 | | Crystal structure of ligand-free human uridine-cytidine kinase 2 | | Descriptor: | Uridine-cytidine kinase 2 | | Authors: | Suzuki, N.N, Koizumi, K, Fukushima, M, Matsuda, A, Inagaki, F. | | Deposit date: | 2003-06-06 | | Release date: | 2004-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase
STRUCTURE, 12, 2004
|
|
8BC1
 
 | | Cryo-EM Structure of Ca2+-bound mTMEM16F F518A_Q623A mutant in GDN | | Descriptor: | Anoctamin-6,mTMEM16F, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
3SEY
 
 | | Zn-mediated Polymer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form II) | | Descriptor: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
1UG7
 
 | | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806 | | Descriptor: | 2610208M17Rik protein | | Authors: | Li, H, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Shirouzu, M, Terada, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-13 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806
To be Published
|
|
1U29
 
 | | Triglycine variant of the ARNO Pleckstrin Homology Domain in complex with Ins(1,4,5)P3 | | Descriptor: | Cytohesin 2, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE | | Authors: | Cronin, T.C, DiNitto, J.P, Czech, M.P, Lambright, D.G. | | Deposit date: | 2004-07-16 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains
Embo J., 23, 2004
|
|
1U2G
 
 | | transhydrogenase (dI.ADPr)2(dIII.NADPH)1 asymmetric complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-19 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
8B8S
 
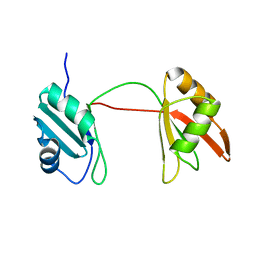 | | Solution structure of tandem RRM1 and RRM2 domains of yeast NPL3 | | Descriptor: | Serine/arginine (SR)-type shuttling mRNA binding protein NPL3 | | Authors: | Kachariya, N, Sattler, M, Keil, P, Strasser, K. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Npl3 functions in mRNP assembly by recruitment of mRNP components to the transcription site and their transfer onto the mRNA.
Nucleic Acids Res., 51, 2023
|
|
8AYJ
 
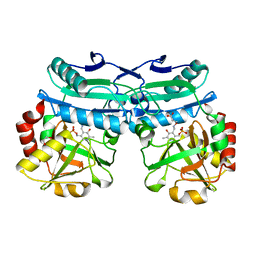 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiens complexed with 3-aminooxypropionic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
