1HM8
 
 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, GLMU, BOUND TO ACETYL COENZYME A | | Descriptor: | ACETYL COENZYME *A, CALCIUM ION, UDP-N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE | | Authors: | Sulzenbacher, G, Gal, L, Peneff, C, Fassy, F, Bourne, Y. | | Deposit date: | 2000-12-05 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture.
J.Biol.Chem., 276, 2001
|
|
2OQJ
 
 | | Crystal structure analysis of Fab 2G12 in complex with peptide 2G12.1 | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, peptide 2G12.1 (ACPPSHVLDMRSGTCLAAEGK) | | Authors: | Calarese, D.A, Stanfield, R.L, Menendez, A, Scott, J.K, Wilson, I.A. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120.
Faseb J., 22, 2008
|
|
5T19
 
 | | Structure of PTP1B complexed with N-(3'-(1,1-dioxido-4-oxo-1,2,5-thiadiazolidin-2-yl)-4'-methyl-[1,1'-biphenyl]-4-yl)acetamide | | Descriptor: | 5-[4-methyl-4'-(methylamino)[1,1'-biphenyl]-3-yl]-1lambda~6~,2,5-thiadiazolidine-1,1,3-trione, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Laciak, A.R, Tanner, J.J. | | Deposit date: | 2016-08-18 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1001 Å) | | Cite: | Covalent Allosteric Inactivation of Protein Tyrosine Phosphatase 1B (PTP1B) by an Inhibitor-Electrophile Conjugate.
Biochemistry, 56, 2017
|
|
1HXJ
 
 | |
1HM6
 
 | |
2BMJ
 
 | | GTPase like domain of Centaurin Gamma 1 (Human) | | Descriptor: | CENTAURIN GAMMA 1 | | Authors: | Yang, X, Elkins, J.M, Soundararajan, M, Arrowsmith, C, Edwards, A, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-14 | | Release date: | 2005-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Centaurin Gamma-1 Gtpase-Like Domain Functions as an Ntpase.
Biochem.J., 401, 2007
|
|
1HN2
 
 | | CRYSTAL STRUCTURE OF BOVINE OBP COMPLEXED WITH AMINOANTHRACENE | | Descriptor: | (3R)-oct-1-en-3-ol, ANTHRACEN-1-YLAMINE, ODORANT-BINDING PROTEIN | | Authors: | Vincent, F, Spinelli, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The insect attractant 1-octen-3-ol is the natural ligand of bovine odorant-binding protein.
J.Biol.Chem., 276, 2001
|
|
1XQ0
 
 | | Structure of human carbonic anhydrase II with 4-[(3-bromo-4-O-sulfamoylbenzyl)(4-cyanophenyl)amino]-4H-[1,2,4]-triazole | | Descriptor: | 2-BROMO-4-{[(4-CYANOPHENYL)(4H-1,2,4-TRIAZOL-4-YL)AMINO]METHYL}PHENYL SULFAMATE, Carbonic anhydrase II, ZINC ION | | Authors: | Lloyd, M.D, Thiyagarajan, N, Ho, Y.T, Woo, L.W.L, Sutcliffe, O.B, Purohit, A, Reed, M.J, Acharya, K.R, Potter, B.V.L. | | Deposit date: | 2004-10-11 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | First Crystal Structures of Human Carbonic Anhydrase II in Complex with Dual Aromatase-Steroid Sulfatase Inhibitors(,)
Biochemistry, 44, 2005
|
|
4MWE
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[5-chloro-3-(prop-2-en-1-yl)-2-(prop-2-en-1-yloxy)benzyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1475) | | Descriptor: | 1-(3-{[5-chloro-3-(prop-2-en-1-yl)-2-(prop-2-en-1-yloxy)benzyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
2HXL
 
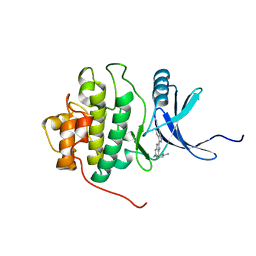 | | crystal structure of Chek1 in complex with inhibitor 1 | | Descriptor: | 3-(5-{[4-(AMINOMETHYL)PIPERIDIN-1-YL]METHYL}-1H-INDOL-2-YL)-1H-INDAZOLE-6-CARBONITRILE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of 6-substituted indolylquinolinones as potent Chek1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3QKG
 
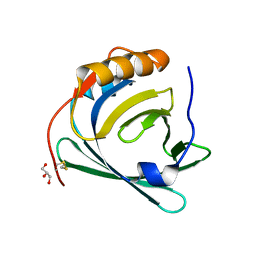 | |
3ALP
 
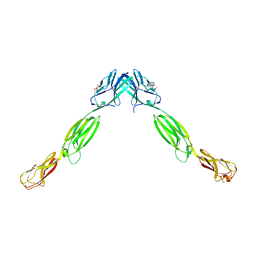 | | Cell adhesion protein | | Descriptor: | CITRIC ACID, HEXANE-1,6-DIOL, Poliovirus receptor-related protein 1 | | Authors: | Narita, H, Nakagawa, A, Suzuki, M. | | Deposit date: | 2010-08-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal Structure of the cis-Dimer of Nectin-1: implications for the architecture of cell-cell junctions
J.Biol.Chem., 286, 2011
|
|
2OWN
 
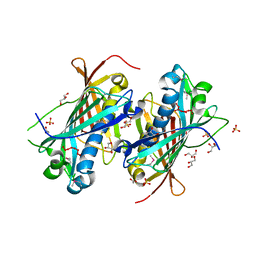 | |
3MB5
 
 | |
4HW4
 
 | |
3PZH
 
 | |
3ZLK
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, N-(6-AMINO-1-BENZYL-2,4-DIOXO-1,2,3,4-TETRAHYDROPYRIMIDIN-5-YL)BENZENESULFONAMIDE | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2013-02-01 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
1YNA
 
 | | ENDO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.0 | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Gruber, K, Kratky, C. | | Deposit date: | 1996-08-22 | | Release date: | 1997-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Thermophilic xylanase from Thermomyces lanuginosus: high-resolution X-ray structure and modeling studies.
Biochemistry, 37, 1998
|
|
3M7O
 
 | | Crystal structure of mouse MD-1 in complex with phosphatidylcholine | | Descriptor: | (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, ... | | Authors: | Harada, H, Ohto, U, Satow, Y. | | Deposit date: | 2010-03-17 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of mouse MD-1 with endogenous phospholipid bound in its cavity
J.Mol.Biol., 400, 2010
|
|
1V08
 
 | | Crystal structure of the Zea maze beta-glucosidase-1 in complex with gluco-tetrazole | | Descriptor: | BETA-GLUCOSIDASE, NOJIRIMYCINE TETRAZOLE | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-25 | | Release date: | 2004-05-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
3UF5
 
 | | Crystal structure of the mouse Colony-Stimulating Factor 1 (mCSF-1) cytokine | | Descriptor: | CALCIUM ION, Macrophage colony-stimulating factor 1 | | Authors: | Elegheert, J, Bracke, N, Bekaert, A, Savvides, S.N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric competitive inactivation of hematopoietic CSF-1 signaling by the viral decoy receptor BARF1
Nat.Struct.Mol.Biol., 19, 2012
|
|
1HJB
 
 | |
3UH8
 
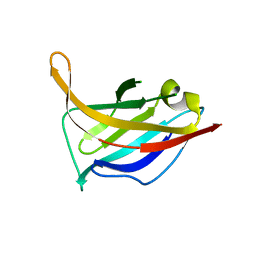 | | N-terminal domain of phage TP901-1 ORF48 | | Descriptor: | ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V.I, van Sinderen, D, Cambillau, C. | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GX6
 
 | |
4GX4
 
 | |
