3LH5
 
 | |
2ROD
 
 | | Solution Structure of MCL-1 Complexed with NoxaA | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, Noxa | | Authors: | Day, C.L, Smits, C, Fan, F.C, Lee, E.F, Fairlie, W.D, Hinds, M.G. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the BH3 Domains from the p53-Inducible BH3-Only Proteins Noxa and Puma in Complex with Mcl-1
J.Mol.Biol., 380, 2008
|
|
4EXY
 
 | |
1DGI
 
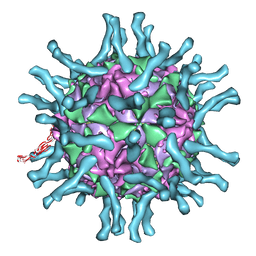 | | Cryo-EM structure of human poliovirus(serotype 1)complexed with three domain CD155 | | Descriptor: | POLIOVIRUS RECEPTOR, VP1, VP2, ... | | Authors: | He, Y, Bowman, V.D, Mueller, S, Bator, C.M, Bella, J, Peng, X, Baker, T.S, Wimmer, E, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 1999-11-24 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (22 Å) | | Cite: | Interaction of the poliovirus receptor with poliovirus.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1H6I
 
 | |
2DCK
 
 | | A tetragonal-lattice structure of alkaliphilic XynJ from Bacillus sp. 41M-1 | | Descriptor: | CALCIUM ION, GLYCEROL, xylanase J | | Authors: | Fibriansah, G, Ihsanawati, Tanaka, N, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-01-07 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1
To be Published
|
|
5XBZ
 
 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminaripentaose | | Descriptor: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Yang, S, Qin, Z, Zhou, P, Yan, Q, Jiang, Z. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
1OD0
 
 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-02-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
3VR4
 
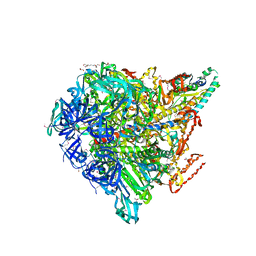 | | Crystal structure of Enterococcus hirae V1-ATPase [eV1] | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saijo, S, Arai, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
2ITB
 
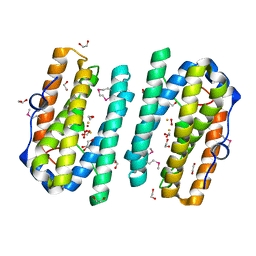 | |
3L5P
 
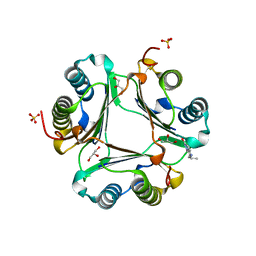 | |
1DJW
 
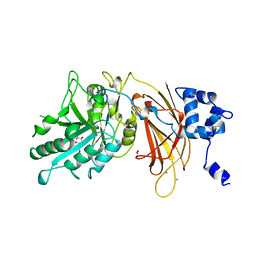 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-2-METHYLENE-1,2-CYCLIC-MONOPHOSPHONATE | | Descriptor: | ACETATE ION, CALCIUM ION, INOSITOL-2-METHYLENE-1,2-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
4ASJ
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, N-(6-amino-1-benzyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-N-methylbenzenesulfonamide, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
1GPU
 
 | | Transketolase complex with reaction intermediate | | Descriptor: | 2-[3-[(4-AMINO-2-METHYL-5-PYRIMIDINYL)METHYL]-2-(1,2-DIHYDROXYETHYL)-4-METHYL-1,3-THIAZOL-3-IUM-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, CALCIUM ION, TRANSKETOLASE 1 | | Authors: | Fiedler, E, Thorell, S, Sandalova, T, Koenig, S, Schneider, G. | | Deposit date: | 2001-11-09 | | Release date: | 2002-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Snapshot of a Key Intermediate in Enzymatic Thiamin Catalysis: Crystal Structure of the Alpha-Carbanion of (Alpha,Beta-Dihydroxyethyl)-Thiamin Diphosphate in the Active Site of Transketolase from Saccharomyces Cerevisiae.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1U30
 
 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors, structure containing maltosyl-alpha (1,4)-D-gluconhydroximo-1,5-lactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
4AS4
 
 | | Structure of human inositol monophosphatase 1 | | Descriptor: | GLYCEROL, INOSITOL MONOPHOSPHATASE 1, MAGNESIUM ION, ... | | Authors: | Singh, N, Knight, M, Halliday, A.C, Lack, N.A, Lowe, E.D, Churchill, G.C. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cloning, Expression, Purification, Crystallization and X-Ray Analysis of Inositol Monophosphatase from Mus Musculus and Homo Sapiens.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4J0L
 
 | | Crystal Structure of a Family GH19 Chitinase (W72A/E67Q mutant) from rye seeds in complex with two (GlcNAc)4 molecules | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic endochitinase C, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2013-01-31 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complete subsite mapping of a "loopful" GH19 chitinase from rye seeds based on its crystal structure
Febs Lett., 587, 2013
|
|
1H0A
 
 | | Epsin ENTH bound to Ins(1,4,5)P3 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, EPSIN | | Authors: | Ford, M.G.J, McMahon, H.T, Evans, P.R. | | Deposit date: | 2002-06-12 | | Release date: | 2002-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Curvature of Clathrin-Coated Pits Driven by Epsin
Nature, 419, 2002
|
|
2MXA
 
 | | Solution structure of the NDH-1 complex subunit CupS from Thermosynechococcus elongatus | | Descriptor: | NDH-1 complex sensory subunit CupS | | Authors: | Korste, A, Wulfhorst, H, Ikegami, T, Nowaczyk, M.M, Stoll, R. | | Deposit date: | 2014-12-17 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the NDH-1 complex subunit CupS from Thermosynechococcus elongatus.
Biochim.Biophys.Acta, 1847, 2015
|
|
3OZ9
 
 | |
4OQ4
 
 | | Crystal Structure of E18A Human DJ-1 | | Descriptor: | Protein DJ-1, SODIUM ION | | Authors: | Prahlad, J, Hauser, D.N, Milkovic, N.M, Cookson, M.R, Wilson, M.A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Use of cysteine-reactive cross-linkers to probe conformational flexibility of human DJ-1 demonstrates that Glu18 mutations are dimers.
J Neurochem, 130, 2014
|
|
2ZNV
 
 | | Crystal structure of human AMSH-LP DUB domain in complex with Lys63-linked ubiquitin dimer | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, Ubiquitin, ... | | Authors: | Sato, Y, Azusa, Y, Yamagata, A, Mimura, H, Wang, X, Yamashita, M, Ookata, K, Nureki, O, Iwai, K, Komada, M, Fukai, S. | | Deposit date: | 2008-05-01 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains
Nature, 455, 2008
|
|
1HHO
 
 | | STRUCTURE OF HUMAN OXYHAEMOGLOBIN AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | HEMOGLOBIN A (OXY) (ALPHA CHAIN), HEMOGLOBIN A (OXY) (BETA CHAIN), OXYGEN MOLECULE, ... | | Authors: | Shaanan, B. | | Deposit date: | 1983-06-10 | | Release date: | 1983-10-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human oxyhaemoglobin at 2.1 A resolution.
J.Mol.Biol., 171, 1983
|
|
2JP8
 
 | | Angiotensin 1-7 | | Descriptor: | Angiotensin-(1-7) | | Authors: | Lula, I, Denadai, A.L, Resende, J.M, de Souza, F.B, de Lima, G.F, Pilo-Veloso, D, Heine, T, Duarte, H.A, Santos, R.A.S, Sinesterra, R.D. | | Deposit date: | 2007-04-27 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Study of angiotensin-(1-7) vasoactive peptide and its beta-cyclodextrin inclusion complexes: complete sequence-specific NMR assignments and structural studies
Peptides, 28, 2007
|
|
3VQQ
 
 | | HIV-1 integrase core domain in complex with 2,1,3-benzothiadiazol-4-amine | | Descriptor: | 2,1,3-benzothiadiazol-4-amine, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
