1DC0
 
 | |
7LXD
 
 | | Structure of yeast DNA Polymerase Zeta (apo) | | Descriptor: | DNA polymerase delta small subunit, DNA polymerase delta subunit 3, DNA polymerase zeta catalytic subunit, ... | | Authors: | Truong, C.D, Craig, T.A, Cui, G, Botuyan, M.V, Serkasevich, R.A, Chan, K.-Y, Mer, G, Chiu, P.-L, Kumar, R. | | Deposit date: | 2021-03-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Cryo-EM reveals conformational flexibility in apo DNA polymerase zeta.
J.Biol.Chem., 297, 2021
|
|
5KIF
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*GP*AP*T)-3'), DNA/RNA (5'-D(*AP*TP*CP*C)-R(P*G)-D(P*GP*TP*AP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
6M3L
 
 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(nonspecific) complex | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*CP*GP*AP*TP*TP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*GP*CP*G)-3'), RE_R_Pab1 domain-containing protein | | Authors: | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
5KGV
 
 | |
5KI4
 
 | |
6UEQ
 
 | | Structure of TBP bound to C-C mismatch containing TATA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*CP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*C)-3'), SULFATE ION, ... | | Authors: | Schumacher, M.A, Al-Hashimi, H. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
5KI7
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*GP*AP*T)-3'), DNA/RNA (5'-D(*AP*TP*CP*C)-R(P*G)-D(P*GP*TP*AP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KI5
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*CP*CP*TP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KIE
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*TP*CP*CP*AP*T)-3'), DNA/RNA (5'-D(*AP*TP*GP*GP*A)-R(P*G)-D(P*CP*TP*C)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KIH
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*CP*CP*TP*AP*A)-3'), DNA/RNA (5'-D(*TP*TP*AP*G)-R(P*G)-D(P*CP*CP*TP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
5KIB
 
 | | Structural impact of single ribonucleotides in DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*CP*CP*TP*AP*A)-3'), DNA/RNA (5'-D(*TP*TP*AP*G)-R(P*G)-D(P*CP*CP*TP*G)-3') | | Authors: | Evich, M, Spring-Connell, A.M, Storici, F, Germann, M.W. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Impact of Single Ribonucleotide Residues in DNA.
Chembiochem, 17, 2016
|
|
7TQQ
 
 | | Structure of human TREX1-DNA complex | | Descriptor: | DNA (5'-D(P*TP*TP*GP*GP*CP*CP*GP*GP*CP*CP*AP*TP*C)-3'), Three-prime repair exonuclease 1 | | Authors: | Zhou, W, Richmond-Buccola, D, Kranzusch, P.J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of human TREX1 DNA degradation and autoimmune disease.
Nat Commun, 13, 2022
|
|
6HMS
 
 | | Cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex with DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*TP*C)-3'), DNA (5'-D(P*TP*GP*AP*CP*GP*CP*GP*GP*CP*CP*CP*GP*TP*CP*TP*C)-3'), DNA polymerase II large subunit,DNA polymerase II large subunit, ... | | Authors: | Raia, P, Carroni, M, Sauguet, L. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases.
PLoS Biol., 17, 2019
|
|
5KUB
 
 | |
7JSG
 
 | | Adeno-Associated Virus 2 Rep68 HD-Heptamer-ssDNA with ATPgS | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
7JSE
 
 | | Adeno-Associated Virus Origin Binding Domain in complex with ssDNA | | Descriptor: | DNA (5'-D(*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
1AAY
 
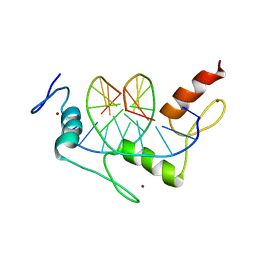 | | ZIF268 ZINC FINGER-DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*GP*CP*CP*CP*AP*CP*GP*C)-3'), PROTEIN (ZIF268 ZINC FINGER PEPTIDE), ... | | Authors: | Elrod-Erickson, M, Rould, M.A, Pabo, C.O. | | Deposit date: | 1997-01-18 | | Release date: | 1997-04-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions.
Structure, 4, 1996
|
|
7KZV
 
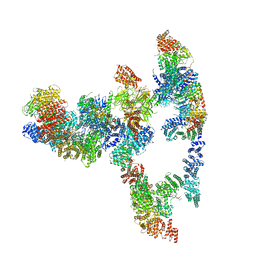 | |
7KZT
 
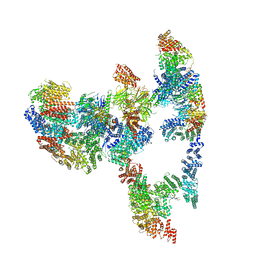 | |
5T2W
 
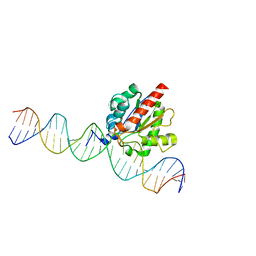 | |
1BF5
 
 | | TYROSINE PHOSPHORYLATED STAT-1/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*G P*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*T P*G)-3'), SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 1-ALPHA/BETA | | Authors: | Kuriyan, J, Zhao, Y, Chen, X, Vinkemeier, U, Jeruzalmi, D, Darnell Jr, J.E. | | Deposit date: | 1998-05-27 | | Release date: | 1998-08-12 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA.
Cell(Cambridge,Mass.), 93, 1998
|
|
3TEK
 
 | | ThermoDBP: a non-canonical single-stranded DNA binding protein with a novel structure and mechanism | | Descriptor: | ThermoDBP-single stranded DNA binding protein | | Authors: | White, M.F, Paytubi, S, Liu, H, Graham, S, McMahon, S.A, Naismith, J.H. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Displacement of the canonical single-stranded DNA-binding protein in the Thermoproteales.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5K5I
 
 | |
5K5L
 
 | |
