5U0B
 
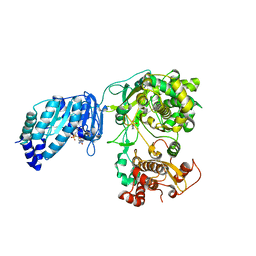 | | Structure of full-length Zika virus NS5 | | Descriptor: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Zhao, B, Du, F. | | Deposit date: | 2016-11-23 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of the Zika virus full-length NS5 protein.
Nat Commun, 8, 2017
|
|
5U19
 
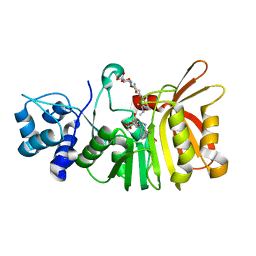 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with (1R,2S,3S,4R,6R)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, MAGNESIUM ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
7VBV
 
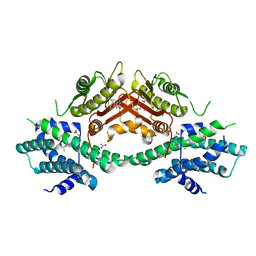 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 7 | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Fujishima, A, Adachi, T. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-based drug design of novel and highly potent pyruvate dehydrogenase kinase inhibitors.
Bioorg.Med.Chem., 52, 2021
|
|
7Z0P
 
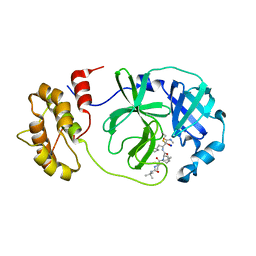 | | SARS-COV2 Main Protease in complex with inhibitor MG-131 | | Descriptor: | (1~{R},2~{S},5~{S})-3-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-~{N}-[(2~{S},3~{R})-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, SODIUM ION | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-02-23 | | Release date: | 2022-04-27 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
8R3D
 
 | | Crystal structure of Pent only at pH 8.8 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Beta propeller | | Authors: | Flood, R.J, Crowley, P.B. | | Deposit date: | 2023-11-08 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Multivalent Calixarene Complexation of a Designed Pentameric Lectin.
Biomacromolecules, 25, 2024
|
|
6MZQ
 
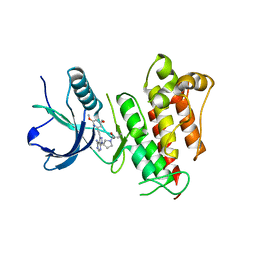 | | TAS-120 in reversible binding mode with FGFR1 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Fibroblast growth factor receptor 1 | | Authors: | Kalyukina, M, Squire, C.J. | | Deposit date: | 2018-11-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TAS-120 Cancer Target Binding: Defining Reactivity and Revealing the First Fibroblast Growth Factor Receptor 1 (FGFR1) Irreversible Structure.
ChemMedChem, 14, 2019
|
|
5CH3
 
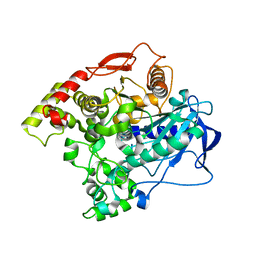 | | E3 alpha-esterase-7 carboxylesterase | | Descriptor: | Carboxylic ester hydrolase | | Authors: | Correy, G, Mabbitt, P, Jackson, C.J. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography.
Structure, 24, 2016
|
|
6MZW
 
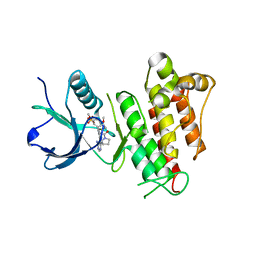 | | TAS-120 covalent complex with FGFR1 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Kalyukina, M, Squire, C.J. | | Deposit date: | 2018-11-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | TAS-120 Cancer Target Binding: Defining Reactivity and Revealing the First Fibroblast Growth Factor Receptor 1 (FGFR1) Irreversible Structure.
ChemMedChem, 14, 2019
|
|
8RYS
 
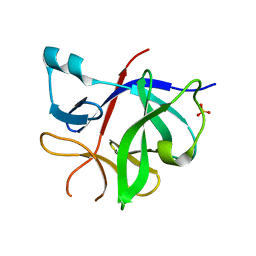 | | Human IL-1beta, unliganded | | Descriptor: | Interleukin-1 beta, SULFATE ION | | Authors: | Rondeau, J.-M, Lehmann, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Ligandability Assessment of IL-1 beta by Integrated Hit Identification Approaches.
J.Med.Chem., 67, 2024
|
|
5U1I
 
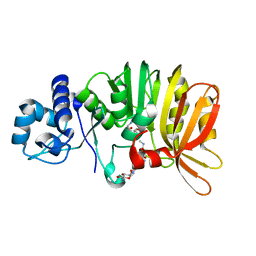 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with the methylated Kanamycin B | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-3-(methylamino)-alpha-D-glucopyranosyl]oxy}-2-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, CALCIUM ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
8UUB
 
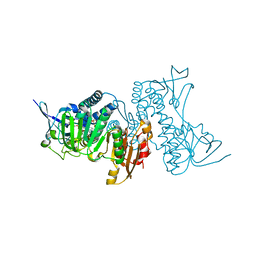 | | Structure of hypothiocyanous acid reductase (Har) from Streptococcus pneumoniae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, Oxidoreductase, ... | | Authors: | Shearer, H.L, Currie, M.J, Dickerhof, N, Dobson, R.C.J. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hypothiocyanous acid reductase is critical for host colonization and infection by Streptococcus pneumoniae.
J.Biol.Chem., 300, 2024
|
|
7YXL
 
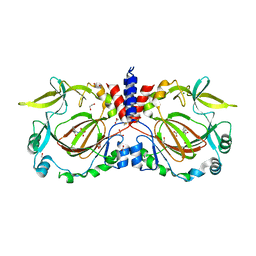 | |
5U1L
 
 | |
6N10
 
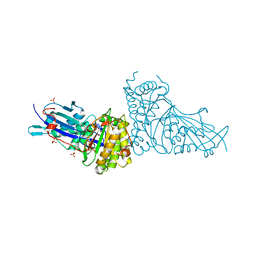 | | Crystal structure of Arabidopsis thaliana mevalonate 5-diphosphate decarboxylase 1 complexed with (R)-MVAPP | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, Diphosphomevalonate decarboxylase MVD1, peroxisomal, ... | | Authors: | Noel, J.P, Thomas, S.T, Louie, G.V. | | Deposit date: | 2018-11-07 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Specificity and Engineering of Mevalonate 5-Phosphate Decarboxylase.
Acs Chem.Biol., 14, 2019
|
|
8UQY
 
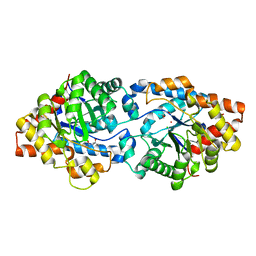 | | Round 18 Arylesterase Variant of Phosphotriesterase Bound to Europium(III) Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, EUROPIUM (III) ION, ... | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6N1A
 
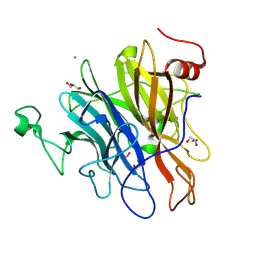 | | Crystal structure of an N-acetylgalactosamine deacetylase from F. plautii | | Descriptor: | CALCIUM ION, Carbohydrate-binding protein, GLYCEROL, ... | | Authors: | Sim, L, Rahfeld, P, Withers, S.G. | | Deposit date: | 2018-11-08 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An enzymatic pathway in the human gut microbiome that converts A to universal O type blood.
Nat Microbiol, 4, 2019
|
|
5CO2
 
 | |
8UUV
 
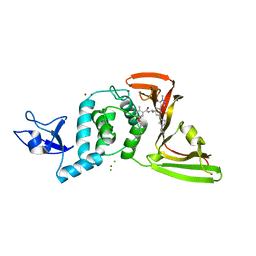 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun12197 | | Descriptor: | CHLORIDE ION, N-{(1R)-1-[(3P,5P)-3-(1-cyclopropyl-1H-pyrazol-4-yl)-5-(1-methyl-1H-pyrazol-4-yl)phenyl]ethyl}-5-[2-(dimethylamino)ethoxy]-2-methylbenzamide, Papain-like protease nsp3, ... | | Authors: | Ansari, A, Tan, B, Arnold, E, Ruiz, F.X, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
7Z0N
 
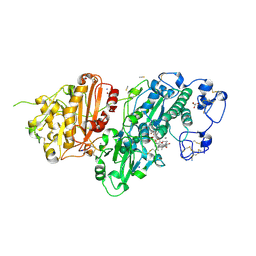 | | Structure-Based Design of a Novel Class of Autotaxin Inhibitors Based on Endogenous Allosteric Modulators | | Descriptor: | CALCIUM ION, GLYCEROL, IODIDE ION, ... | | Authors: | Salgado-Polo, F, Clark, J.M, Macdonald, S.J.F, Barrett, T.N, Perrakis, A, Jamieson, A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of a Novel Class of Autotaxin Inhibitors Based on Endogenous Allosteric Modulators.
J.Med.Chem., 65, 2022
|
|
8P5R
 
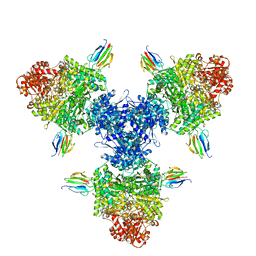 | | Crystal structure of full-length, homohexameric 2-oxoglutarate dehydrogenase KGD from Mycobacterium smegmatis in complex with GarA | | Descriptor: | CALCIUM ION, Glycogen accumulation regulator GarA, MAGNESIUM ION, ... | | Authors: | Wagner, T, Mechaly, A.M, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.562 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8R3B
 
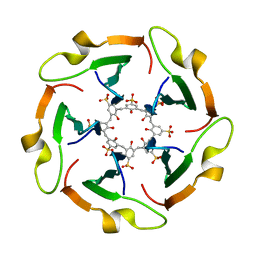 | |
8UD0
 
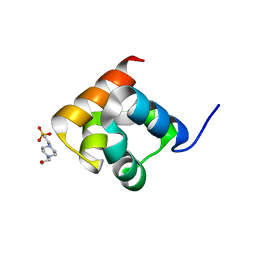 | |
7PQ3
 
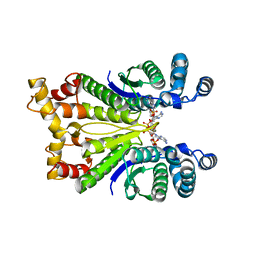 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in complex with its post-catalytic reaction product | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
7PQ2
 
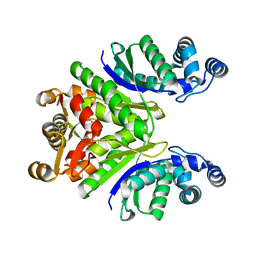 | | Crystal Structure of the Ring Nuclease 0811 from Sulfolobus islandicus (Sis0811) in its apo form | | Descriptor: | CRISPR-associated protein, APE2256 family, CRISPR Ring Nuclease | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
5CP1
 
 | |
