6G1V
 
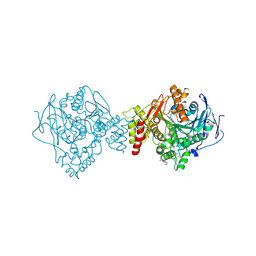 | | Crystal structure of Torpedo Californica acetylcholinesterase in complex with 12-Amino-3-chloro-6,7,10,11-tetrahydro-5,9-dimethyl-7,11-methanocycloocta[b]quinolin-5-ium | | Descriptor: | 12-Amino-3-chloro-6,7,10,11-tetrahydro-5,9-dimethyl-7,11-methanocycloocta[b]quinolin-5-ium, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Coquelle, N, Colletier, J.P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Increasing Polarity in Tacrine and Huprine Derivatives: Potent Anticholinesterase Agents for the Treatment of Myasthenia Gravis.
Molecules, 23, 2018
|
|
8GBU
 
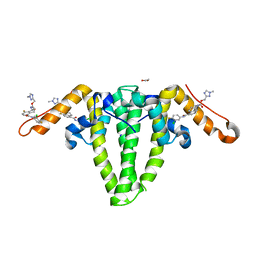 | | Hepatitis B capsid Y132A mutant with compound AB-506 | | Descriptor: | (1-methyl-1H-1,2,4-triazol-3-yl)methyl {(1S)-4-[(3-chloro-4-fluorophenyl)carbamoyl]-7-fluoro-2,3-dihydro-1H-inden-1-yl}carbamate, 1,2-ETHANEDIOL, Capsid protein | | Authors: | Horanyi, P.S, Mayclin, S.J. | | Deposit date: | 2023-02-28 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis, and structure-activity relationship of a bicyclic HBV capsid assembly modulator chemotype leading to the identification of clinical candidate AB-506.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
3LJ3
 
 | | PI3-kinase-gamma with a pyrrolopyridine-benzofuran inhibitor | | Descriptor: | (2Z)-4,6-dihydroxy-2-{[1-methyl-4-(4-methylpiperazin-1-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl]methylidene}-1-benzofuran-3(2H)-one, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Bard, J, Svenson, K. | | Deposit date: | 2010-01-25 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery and optimization of 2-(4-substituted-pyrrolo[2,3-b]pyridin-3-yl)methylene-4-hydroxybenzofuran-3(2H)-ones as potent and selective ATP-competitive inhibitors of the mammalian target of rapamycin (mTOR).
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7ORA
 
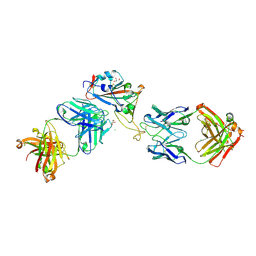 | | Crystal structure of the T478K mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-45 and COVOX-253 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-253 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7SBG
 
 | |
8Q60
 
 | | Racemic crystal structure of a synthetic DNA Hairpin with diquinoline linker. | | Descriptor: | 4-[8-(aminomethyl)-2-[[5-(3-oxidanylpropyl)quinolin-8-yl]carbamoyl]quinolin-4-yl]butyl dihydrogen phosphite, 5'-D(*GP*TP*TP*TP*TP*G)-3', DNA (5'-D(CP*AP*AP*AP*AP*C)-3'), ... | | Authors: | Mandal, P.K, Loos, M, Xu, F, Huc, I. | | Deposit date: | 2023-08-10 | | Release date: | 2025-02-19 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interfacing B-DNA and DNA Mimic Foldamers.
Angew.Chem.Int.Ed.Engl., 2025
|
|
7ZJQ
 
 | |
6MMD
 
 | | Photoactive Yellow Protein with 3,5-dichlorotyrosine substituted at position 42 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-30 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.228 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
5L3J
 
 | | ESCHERICHIA COLI DNA GYRASE B IN COMPLEX WITH BENZOTHIAZOLE-BASED INHIBITOR | | Descriptor: | 2-[[2-[[4,5-bis(bromanyl)-1~{H}-pyrrol-2-yl]carbonylamino]-1,3-benzothiazol-6-yl]amino]-2-oxidanylidene-ethanoic acid, DNA gyrase subunit B, IODIDE ION | | Authors: | Gjorgjieva, M, Tomasic, T, Barancokova, M, Katsamakas, S, Ilas, J, Montalvao, S, Tammella, P, Peterlin Masic, L, Kikelj, D. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Discovery of Benzothiazole Scaffold-Based DNA Gyrase B Inhibitors.
J.Med.Chem., 59, 2016
|
|
8EF2
 
 | |
6NSL
 
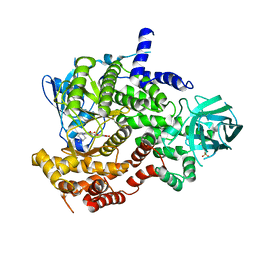
 | | CRYSTAL STRUCTURE OF TYROSINE KINASE 2 JH2 (PSEUDO KINASE DOMAIN) COMPLEXED WITH Compound-6c AKA 6-((1-(4-CYANOPHENY L)-2-OXO-1,2-DIHYDRO-3-PYRIDINYL)AMINO)-N-CYCLOPROPYL-8-(M ETHYLAMINO)IMIDAZO[1,2-B]PYRIDAZINE-3-CARBOXAMIDE | | Descriptor: | 6-{[1-(4-cyanophenyl)-2-oxo-1,2-dihydropyridin-3-yl]amino}-N-cyclopropyl-8-(methylamino)imidazo[1,2-b]pyridazine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Muckelbauer, J.M, Khan, J.A. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of Imidazo[1,2-b]pyridazine Derivatives as Potent, Selective, and Orally Active Tyk2 JH2 Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
7U4N
 
 | | Crystal structure of human GPX4-U46C in complex with RSL3 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, methyl (1S,3R)-2-(chloroacetyl)-1-[4-(methoxycarbonyl)phenyl]-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7USJ
 
 | | BRD4-BD2 in complex with SF2523 | | Descriptor: | 3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-(morpholin-4-yl)-7H-thieno[3,2-b]pyran-7-one, Bromodomain-containing protein 4 | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2022-04-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Targeting BRD4 and PI3K signaling pathways for the treatment of medulloblastoma.
J Control Release, 354, 2023
|
|
4YOA
 
 | | Crsystal structure HIV-1 Protease MDR769 L33F Complexed with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 Protease | | Authors: | Kuiper, B.D, Keusch, B, Dewdney, T.G, Chordia, P, Brunzelle, J.S, Ross, K, Kovari, I.A, MacArthur, R, Salimnia, H, Kovari, L.C. | | Deposit date: | 2015-03-11 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | The L33F darunavir resistance mutation acts as a molecular anchor reducing the flexibility of the HIV-1 protease 30s and 80s loops.
Biochem Biophys Rep, 2, 2015
|
|
5M3A
 
 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-6-(1-methyl-5-phenoxy-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
5IFA
 
 | |
5FNU
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(7-methoxy-1-methyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)phenyl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
6YJJ
 
 | |
9GAX
 
 | | TEAD1 in complex with a reversible inhibitor N-[(1S)-2-hydroxy-1-(1-methyl-1H-pyrazol-3-yl)ethyl]-2-methyl-8-[4-(trifluoromethyl)phenyl]-2H,8H-pyrazolo[3,4-b]indole-5-carboxamide | | Descriptor: | 2-methyl-~{N}-[(1~{S})-1-(1-methylpyrazol-3-yl)-2-oxidanyl-ethyl]-4-[4-(trifluoromethyl)phenyl]pyrazolo[3,4-b]indole-7-carboxamide, GLYCEROL, SULFATE ION, ... | | Authors: | Musil, D, Freire, F. | | Deposit date: | 2024-07-29 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | MoA Studies of the TEAD P-Site Binding Ligand MSC-4106 and Its Optimization to TEAD1-Selective Amide M3686.
J.Med.Chem., 68, 2025
|
|
8QH1
 
 | |
8QH0
 
 | | Crystal structure of the SARS-CoV-2 RBD with the antibody Cv2.3194 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cv2.3194 Heavy chain, GLYCEROL, ... | | Authors: | Fernandez, I, Rey, F.A. | | Deposit date: | 2023-09-06 | | Release date: | 2024-06-19 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Broad sarbecovirus neutralization by combined memory B cell antibodies to ancestral SARS-CoV-2.
Iscience, 27, 2024
|
|
7NE4
 
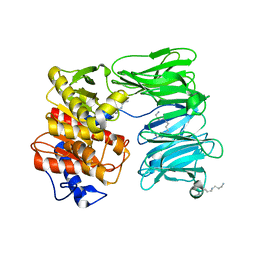 | | E125A mutant of oligopeptidase B from S. proteomaculans with modified hinge region | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | First Crystal Structure of Bacterial Oligopeptidase B in an Intermediate State: The Roles of the Hinge Region Modification and Spermine.
Biology (Basel), 10, 2021
|
|
5ZKB
 
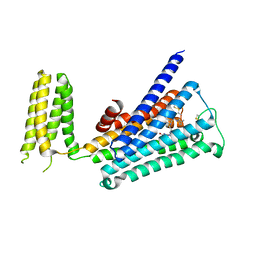 | | Crystal structure of rationally thermostabilized M2 muscarinic acetylcholine receptor bound with AF-DX 384 | | Descriptor: | Muscarinic acetylcholine receptor M2,Apo-cytochrome b562,Muscarinic acetylcholine receptor M2, N-[2-[(2S)-2-[(dipropylamino)methyl]piperidin-1-yl]ethyl]-6-oxidanylidene-5H-pyrido[2,3-b][1,4]benzodiazepine-11-carboxamide | | Authors: | Suno, R, Maeda, S, Yasuda, S, Yamashita, K, Hirata, K, Horita, S, Tawaramoto, M.S, Tsujimoto, H, Murata, T, Kinoshita, M, Yamamoto, M, Kobilka, B.K, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the subtype-selective antagonist binding to the M2muscarinic receptor
Nat. Chem. Biol., 14, 2018
|
|
1APF
 
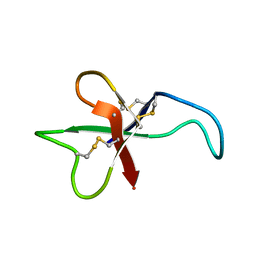 | | ANTHOPLEURIN-B, NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-B | | Authors: | Monks, S.A, Pallaghy, P.K, Scanlon, M.J, Norton, R.S. | | Deposit date: | 1995-05-30 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cardiostimulant polypeptide anthopleurin-B and comparison with anthopleurin-A.
Structure, 3, 1995
|
|
9GGN
 
 | | Cryo-EM structure of KBTBD4 WT-HDAC2 2:2 complex mediated by molecular glue UM171 | | Descriptor: | (1r,4r)-N~1~-[(7P)-2-benzyl-7-(2-methyl-2H-tetrazol-5-yl)-9H-pyrimido[4,5-b]indol-4-yl]cyclohexane-1,4-diamine, Histone deacetylase 2, Isoform 1 of Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Chen, Z, Chi, G, Pike, A.C.W, Montes, B, Bullock, A.N. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mimicry of UM171 and neomorphic cancer mutants co-opts E3 ligase KBTBD4 for HDAC1/2 recruitment.
Nat Commun, 16, 2025
|
|
