4OYS
 
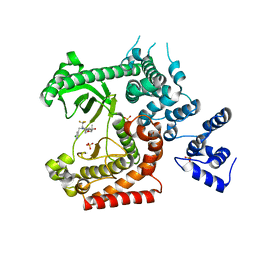 | | CRYSTAL STRUCTURE OF VPS34 IN COMPLEX WITH SAR405. | | Descriptor: | (8S)-9-[(5-chloranylpyridin-3-yl)methyl]-2-[(3R)-3-methylmorpholin-4-yl]-8-(trifluoromethyl)-6,7,8,9a-tetrahydro-3H-pyrimido[1,2-a]pyrimidin-4-one, Phosphatidylinositol 3-kinase catalytic subunit type 3, SULFATE ION | | Authors: | Mathieu, M, Marquette, J.p. | | Deposit date: | 2014-02-13 | | Release date: | 2014-10-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy.
Nat.Chem.Biol., 10, 2014
|
|
3J3Z
 
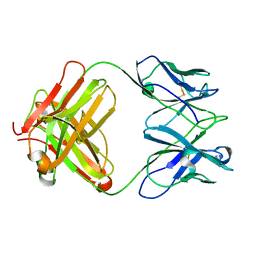 | | Structure of MA28-7 neutralizing antibody Fab fragment from electron cryo-microscopy of enterovirus 71 complexed with a Fab fragment | | Descriptor: | MA28-7 neutralizing antibody heavy chain, MA28-7 neutralizing antibody light chain | | Authors: | Lee, H, Cifuente, J.O, Ashley, R.E, Conway, J.F, Makhov, A.M, Tano, Y, Shimizu, H, Nishimura, Y, Hafenstein, S. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (23.4 Å) | | Cite: | A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with fab from neutralizing antibody.
J.Virol., 87, 2013
|
|
7JXH
 
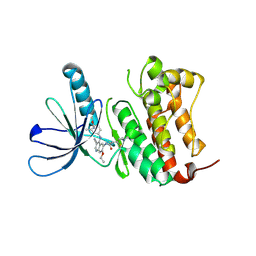 | | HER2 in complex with JBJ-08-178-01 | | Descriptor: | (2E)-N-[3-cyano-7-ethoxy-4-({3-methyl-4-[([1,2,4]triazolo[1,5-a]pyridin-7-yl)oxy]phenyl}amino)quinolin-6-yl]-4-(dimethylamino)but-2-enamide, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | A Novel HER2-Selective Kinase Inhibitor Is Effective in HER2 Mutant and Amplified Non-Small Cell Lung Cancer.
Cancer Res., 82, 2022
|
|
3NE0
 
 | |
7MLE
 
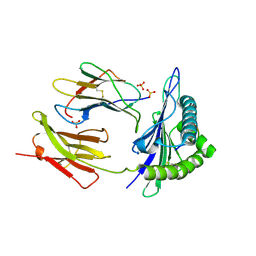 | | Crystal Structure of HLA-A*03:01 in complex with VVRPSVASK, an 9-mer epitope from Influenza B virus | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homologous peptides derived from influenza A, B and C viruses induce variable CD8 + T cell responses with cross-reactive potential.
Clin Transl Immunology, 11, 2022
|
|
5MVH
 
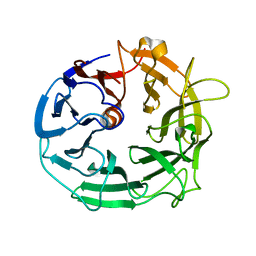 | | Glycoside Hydrolase BACCELL_00856 | | Descriptor: | BACCELL_00856 | | Authors: | Munoz-Munoz, J, Cartmell, A, Terrapon, N, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-01-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual active site location and catalytic apparatus in a glycoside hydrolase family.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6BWD
 
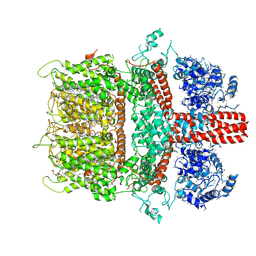 | | 3.7 angstrom cryoEM structure of truncated mouse TRPM7 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, MAGNESIUM ION, Transient receptor potential cation channel subfamily M member 7 | | Authors: | Zhang, J, Li, Z, Duan, J, Li, J, Hulse, R.E, Santa-Cruz, A, Abiria, S.A, Krapivinsky, G, Clapham, D.E. | | Deposit date: | 2017-12-14 | | Release date: | 2018-08-15 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the mammalian TRPM7, a magnesium channel required during embryonic development.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MUL
 
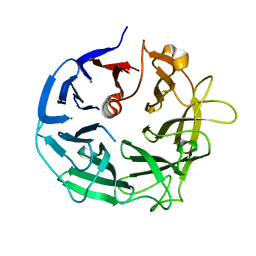 | | Glycoside Hydrolase BT3686 bound to Glucuronic Acid | | Descriptor: | Neuraminidase, beta-D-glucopyranuronic acid | | Authors: | Munoz-Munoz, J, Cartmell, A, Terrapon, N, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Unusual active site location and catalytic apparatus in a glycoside hydrolase family.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2V8I
 
 | |
6ZCE
 
 | | Structure of a yeast ABCE1-bound 43S pre-initiation complex | | Descriptor: | 18S ribosomal RNA (1719-MER), 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-07 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
4LXV
 
 | | Crystal Structure of the Hemagglutinin from a H1N1pdm A/WASHINGTON/5/2011 virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Chang, J.C, Guo, Z, Carney, P.J, Shore, D.A, Donis, R.O, Cox, N.J, Villanueva, J.M, Klimov, A.I, Stevens, J. | | Deposit date: | 2013-07-30 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Stability of Influenza A(H1N1)pdm09 Virus Hemagglutinins.
J.Virol., 88, 2014
|
|
3U6T
 
 | | Crystal structure of the complex of type I Ribosome inactivating protein in complex with Kanamycin at 1.85 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, KANAMYCIN A, ... | | Authors: | Yamini, S, Pandey, S, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-10-13 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the complex of type I Ribosome inactivating protein in complex with Kanamycin at 1.85 A
To be Published
|
|
6CGN
 
 | |
6D0K
 
 | | Crystal structure of a CLC-type fluoride/proton antiporter, E118Q mutant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CK2
 
 | | Insulin analog containing a YB26W mutation | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Rege, N.K, Yee, V.C, Weiss, M.A. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based stabilization of insulin as a therapeutic protein assembly via enhanced aromatic-aromatic interactions.
J. Biol. Chem., 293, 2018
|
|
5UIG
 
 | | Crystal structure of adenosine A2A receptor bound to a novel triazole-carboximidamide antagonist | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-amino-N-[(2-methoxyphenyl)methyl]-2-(3-methylphenyl)-2H-1,2,3-triazole-4-carboximidamide, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Sun, B, Bachhawat, P, Ling-Hon Chu, M, Ceska, T, Sands, Z, Lebon, F, Kobilka, T.S, Kobilka, B. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the adenosine A2A receptor bound to an antagonist reveals a potential allosteric pocket.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KDV
 
 | | IMPa metallopeptidase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Metallopeptidase, SULFATE ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5O49
 
 | | Human FGF in complex with a covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl 3-fluorosulfonylbenzoate | | Authors: | Debreczeni, J, Breed, J, Mukherjee, H, Aquila, B, Kaiser, C, Tentarelli, S, Whitty, A, Grimster, N. | | Deposit date: | 2017-05-26 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | A study of the reactivity of S(VI)-F containing warheads with nucleophilic amino-acid side chains under physiological conditions.
Org. Biomol. Chem., 15, 2017
|
|
5E2Z
 
 | |
6CPF
 
 | | Structure of dephosphorylated Aurora A (122-403) bound to AMPPCP in an active conformation | | Descriptor: | Aurora kinase A, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Otten, R, Zorba, A, Padua, R.A.P, Kern, D. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamics of human protein kinase Aurora A linked to drug selectivity.
Elife, 7, 2018
|
|
5KDW
 
 | | IMPa metallopeptidase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Metallopeptidase, PHOSPHATE ION, ... | | Authors: | Noach, I, Boraston, A.B. | | Deposit date: | 2016-06-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Recognition of protein-linked glycans as a determinant of peptidase activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6RVH
 
 | | NADH-dependent Coenzyme A Disulfide Reductase soaked with Menadione | | Descriptor: | COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, MENADIONE, ... | | Authors: | Koepke, J, Preu, J. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Characterization and X-ray structure of the NADH-dependent coenzyme A disulfide reductase from Thermus thermophilus.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
6RUZ
 
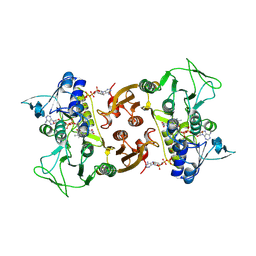 | | NADH-dependent Coenzyme A Disulfide Reductase | | Descriptor: | COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, NADH oxidase | | Authors: | Koepke, J, Preu, J. | | Deposit date: | 2019-05-29 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization and X-ray structure of the NADH-dependent coenzyme A disulfide reductase from Thermus thermophilus.
Biochim Biophys Acta Bioenerg, 1860, 2019
|
|
1NH6
 
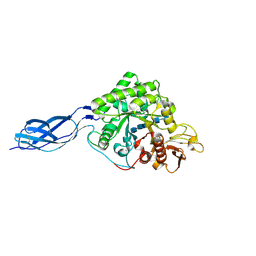 | | Structure of S. marcescens chitinase A, E315L, complex with hexasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, chitinase A | | Authors: | Aronson Jr, N.N, Halloran, B.A, Alexyev, M.F, Amable, L, Madura, J.D, Pasupulati, L, Worth, C, Van Roey, P. | | Deposit date: | 2002-12-18 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Family 18 chitinase-oligosaccharide substrate interaction: subsite preference and anomer selectivity of Serratia marcescens chitinase A.
Biochem.J., 376, 2003
|
|
6DFP
 
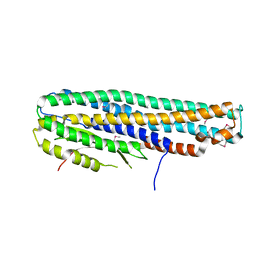 | | Crystal Structure of a Tripartite Toxin Component VCA0883 from Vibrio cholerae | | Descriptor: | VCA0883 | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
