4W5C
 
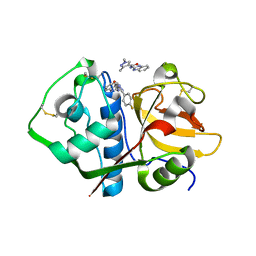 | | Crystal structure analysis of cruzain with three Fragments: 1 (N-(1H-benzimidazol-2-yl)-1,3-dimethyl-pyrazole-4-carboxamide), 6 (2-amino-4,6-difluorobenzothiazole) and 9 (N-(1H-benzimidazol-2-yl)-3-(4-fluorophenyl)-1H-pyrazole-4-carboxamide). | | Descriptor: | 4,6-difluoro-1,3-benzothiazol-2-amine, Cruzipain, N-(1H-benzimidazol-2-yl)-1,3-dimethyl-1H-pyrazole-4-carboxamide, ... | | Authors: | Tochowicz, A, McKerrow, J.H, Craik, C.S. | | Deposit date: | 2014-08-17 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Applying Fragments Based- Drug Design to identify multiple binding modes on cysteine protease.
To Be Published
|
|
4W78
 
 | | Crystal structure of the ChsH1-ChsH2 complex from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Guja, K.E, Yang, M, Sampson, N, Garcia-Diaz, M. | | Deposit date: | 2014-08-21 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | A Distinct MaoC-like Enoyl-CoA Hydratase Architecture Mediates Cholesterol Catabolism in Mycobacterium tuberculosis.
Acs Chem.Biol., 9, 2014
|
|
4W8E
 
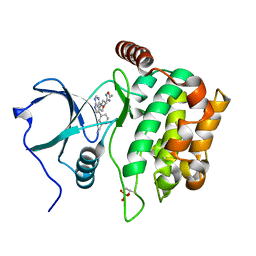 | | Structure of MST3 with a pyrrolopyrimidine inhibitor (PF-06645342) | | Descriptor: | 3-{4-[(2R)-2-(5-methyl-1,2,4-oxadiazol-3-yl)morpholin-4-yl]-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Serine/threonine-protein kinase 24 36 kDa subunit | | Authors: | Jasti, J, Song, X, Griffor, M, Kurumbail, R.G. | | Deposit date: | 2014-08-24 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery and preclinical profiling of 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile (PF-06447475), a highly potent, selective, brain penetrant, and in vivo active LRRK2 kinase inhibitor.
J.Med.Chem., 58, 2015
|
|
4W8K
 
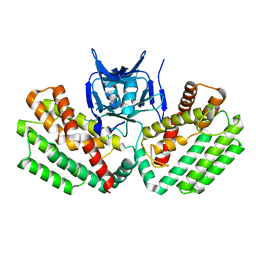 | | Crystal structure of a putative Cas1 enzyme from Vibrio phage ICP1 | | Descriptor: | Cas1 protein, POTASSIUM ION | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyeno, O, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To Be Published
|
|
4V3J
 
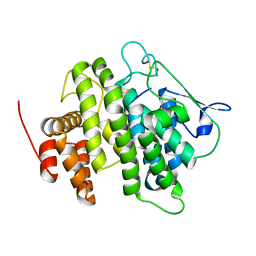 | | Structural and functional characterization of a novel monotreme- specific protein from the milk of the platypus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MONOTREME LACTATING PROTEIN | | Authors: | Kumar, A, Newman, J, Polekina, G, Adams, T.E, Sharp, J.A, Peat, T.S, Nicholas, K.R. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural characterization of a novel monotreme-specific protein with antimicrobial activity from the milk of the platypus.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4V46
 
 | | Crystal structure of the BAFF-BAFF-R complex | | Descriptor: | MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Kim, H.M, Yu, K.S, Lee, M.E, Shin, D.R, Kim, Y.S, Paik, S.G, Yoo, O.J, Lee, H, Lee, J.-O. | | Deposit date: | 2003-03-23 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation
NAT.STRUCT.BIOL., 10, 2003
|
|
4UXS
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
4UY1
 
 | | Novel pyrazole series of group X Secretory Phospholipase A2 (sPLA2-X) inhibitors | | Descriptor: | 5-(2,5-DIMETHYL-3-THIENYL)-1H-PYRAZOLE-3-CARBOXAMIDE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J, Oster, L, Hallberg, K, Bodin, C, Chen, H. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-15 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Pyrazole Series of Group X Secreted Phospholipase A2 Inhibitor (Spla2X) Via Fragment Based Virtual Screening
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4V0X
 
 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-684) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
8XT1
 
 | | Cryo-EM structure of the human 39S mitoribosome with 5uM Tigecycline | | Descriptor: | 16s rRNA, 39S ribosomal protein L22, mitochondrial, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8Y39
 
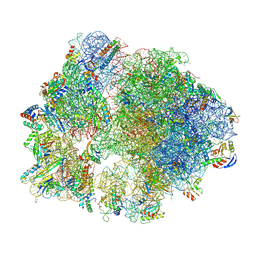 | | cryo-EM structure of Staphylococcus aureus(ATCC 29213) 70S ribosome in complex with MCX-190. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
6WDD
 
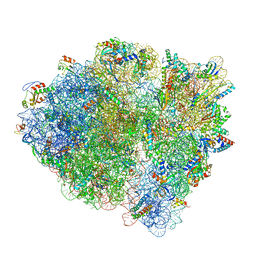 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure V-A) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
2Y82
 
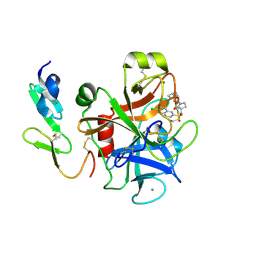 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-((3S)-2-OXO-1-{4-[(2S)-2-PYRROLIDINYL]PHENYL}-3-PYRROLIDINYL)-2-NAPHTHALENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
9ATD
 
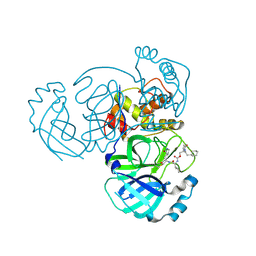 | | Crystal structure of MERS 3CL protease in complex with a ethylcyclohexyl 2-pyrrolidone inhibitor (S-enantiomer) inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(2S)-1-(2-cyclohexylethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-(2-cyclohexylethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
2YGW
 
 | | Crystal structure of human MCD | | Descriptor: | 1,2-ETHANEDIOL, MALONYL-COA DECARBOXYLASE, MITOCHONDRIAL, ... | | Authors: | Vollmar, M, Puranik, S, Krojer, T, Savitsky, P, Allerston, C, Yue, W.W, Chaikuad, A, von Delft, F, Gileadi, O, Kavanagh, K, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Oppermann, U. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Malonyl-Coenzyme a Decarboxylase Provide Insights Into its Catalytic Mechanism and Disease-Causing Mutations.
Structure, 21, 2013
|
|
7B97
 
 | | CooS-V with oxidized hybrid cluster by hydroxylamine for 30 min | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 3,5-dioxa-7-thia-1-thionia-2$l^{2},4$l^{2},6$l^{3},8$l^{2}-tetraferrabicyclo[4.2.0]octane, Carbon monoxide dehydrogenase, ... | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Morphing [4Fe-3S-nO]-Cluster within a Carbon Monoxide Dehydrogenase Scaffold.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6WDM
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Non-cognate Structure V-B2) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
7B9Q
 
 | | The SERp optimized structure of Ribonucleotide reductase from Rhodobacter sphaeroides | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Vitamin B12-dependent ribonucleotide reductase | | Authors: | Loderer, C, Feiler, C, Wilk, P, Kabinger, F. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | HUG Domain Is Responsible for Active Dimer Stabilization in an NrdJd Ribonucleotide Reductase.
Biochemistry, 61, 2022
|
|
8Y7L
 
 | |
6WHM
 
 | | The crystal structure of the 2009/H1N1/California PA endonuclease wild type bound to DNA oligomer TAGC (cleaved TTAGCATT, 5mM overnight DNA soak) | | Descriptor: | DNA (5'-D(P*TP*AP*GP*C)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Webb, T, White, S.W. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
8Y37
 
 | | Cryo-EM structure of Staphylococcus aureus (15B196) 50S ribosome in complex with MCX-190. | | Descriptor: | 23S ribosomal RNA, 5S ribosomal RNA, 7-[4-[3-[[(1~{S},2~{R},5~{R},6~{S},7~{S},8~{R},9~{R},11~{R},13~{R},14~{R})-8-[(2~{S},3~{R},4~{S},6~{R})-4-(dimethylamino)-6-methyl-3-oxidanyl-oxan-2-yl]oxy-2-ethyl-9-methoxy-1,5,7,9,11,13-hexamethyl-4,12,16-tris(oxidanylidene)-3,17-dioxa-15-azabicyclo[12.3.0]heptadecan-6-yl]oxycarbonylamino]propoxy]but-1-ynyl]-1-methyl-4-oxidanylidene-quinoline-3-carboxylic acid, ... | | Authors: | Li, Y, Lu, G, Li, J, Pei, X, Lin, J. | | Deposit date: | 2024-01-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Synthetic macrolides overcoming MLS B K-resistant pathogens.
Cell Discov, 10, 2024
|
|
2YNK
 
 | | Wzi, an Outer Membrane Protein Involved in Group 1 Capsule Assembly in Escherichia coli, is a Carbohydrate Binding Beta-Barrel | | Descriptor: | DODECANE, HEXANE, N-OCTANE, ... | | Authors: | Bushell, S.R, Mainprize, I.L, Wear, M.A, Lou, H, Kong, L, Davis, B, Bayley, H, Whitfield, C, Naismith, J.H. | | Deposit date: | 2012-10-16 | | Release date: | 2013-05-15 | | Last modified: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Wzi is an Outer Membrane Lectin that Underpins Group 1 Capsule Assembly in Escherichia Coli.
Structure, 21, 2013
|
|
9AZJ
 
 | |
8YIF
 
 | | Crystal structure of GH13_30 alpha-glucosidase CmmB in complex with acarviosin | | Descriptor: | Acarviosin, Alpha-glucosidase | | Authors: | Saburi, W, Tagami, T, Yu, J, Ose, T, Yao, M, Mori, H. | | Deposit date: | 2024-02-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism for the substrate specificity of Arthrobacter globiformis M6 alpha-glucosidase CmmB, belonging to glycoside hydrolase family 13 subfamily 30
Food Biosci, 61, 2024
|
|
2YNV
 
 | | Cys221 oxidized, Mono zinc GIM-1 - GIM-1-Ox. Crystal structures of Pseudomonas aeruginosa GIM-1: active site plasticity in metallo-beta- lactamases | | Descriptor: | GIM-1 PROTEIN, MAGNESIUM ION, ZINC ION | | Authors: | Borra, P.S, Samuelsen, O, Spencer, J, Lorentzen, M.S, Leiros, H.-K.S. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-24 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of Pseudomonas Aeruginosa Gim-1: Active-Site Plasticity in Metallo-Beta-Lactamases.
Antimicrob.Agents Chemother., 57, 2013
|
|
