8QCN
 
 | | E.coli IspE in complex with a ligand (2) | | Descriptor: | 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase, ADENOSINE-5'-DIPHOSPHATE, ~{N}-[3-(4-azanyl-2-oxidanylidene-1~{H}-pyrimidin-5-yl)prop-2-ynyl]cyclopropanesulfonamide | | Authors: | Hamid, R. | | Deposit date: | 2023-08-27 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | E.coli IspE in complex with a ligand (2)
To Be Published
|
|
8QCO
 
 | | E.coli IspE in complex with a ligand (3) | | Descriptor: | 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase, ADENOSINE-5'-DIPHOSPHATE, ~{N}-[3-(4-azanyl-2-oxidanylidene-1~{H}-pyrimidin-5-yl)prop-2-ynyl]cyclopentanesulfonamide | | Authors: | Hamid, R. | | Deposit date: | 2023-08-28 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | E.coli IspE in complex with a ligand (3)
To Be Published
|
|
2Y2T
 
 | |
1BQ2
 
 | | E. COLI THYMIDYLATE SYNTHASE MUTANT N177A | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Reyes, C.L, Sage, C.R, Rutenber, E.E, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inactivity of N229A thymidylate synthase due to water-mediated effects: isolating a late stage in methyl transfer.
J.Mol.Biol., 284, 1998
|
|
5VPN
 
 | | E. coli Quinol fumarate reductase FrdA E245Q mutation | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Starbird, C.A, Maklashina, E, Sharma, P, Qualls-Histed, S, Cecchini, G, Iverson, T.M. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.2232 Å) | | Cite: | Structural and biochemical analyses reveal insights into covalent flavinylation of the Escherichia coli Complex II homolog quinol:fumarate reductase.
J. Biol. Chem., 292, 2017
|
|
2PJJ
 
 | |
1KMJ
 
 | |
4N99
 
 | | E. coli sliding clamp in complex with 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-7-carboxylic acid | | Descriptor: | 6-chloro-2,3,4,9-tetrahydro-1H-carbazole-7-carboxylic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N94
 
 | | E. coli sliding clamp in complex with 3,4-difluorobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 3,4-difluorobenzamide, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
6HJK
 
 | | Crystal Structure of Aurora-A L210C catalytic domain in complex with ASDO2 | | Descriptor: | (~{E})-~{N}-[4-(4-azanyl-1-propan-2-yl-pyrazolo[3,4-d]pyrimidin-3-yl)phenyl]-4-[4-fluoranyl-3-(trifluoromethyl)phenyl]-4-oxidanylidene-but-2-enamide, Aurora kinase A, CHLORIDE ION, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2018-09-04 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Type II Kinase Inhibitors Targeting Cys-Gatekeeper Kinases Display Orthogonality with Wild Type and Ala/Gly-Gatekeeper Kinases.
ACS Chem. Biol., 13, 2018
|
|
1KMK
 
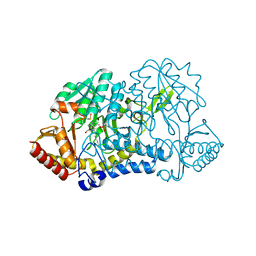 | |
4N98
 
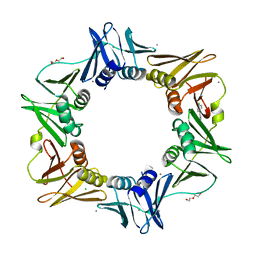 | |
4PNC
 
 | | E. COLI METHIONINE AMINOPEPTIDASE IN COMPLEX WITH INHIBITOR 7-METHOXY-2-METHYLEN-3,4-DIHYDRONAPHTHALEN-1(2H)-ONE | | Descriptor: | (2S)-7-methoxy-2-methyl-3,4-dihydronaphthalen-1(2H)-one, COBALT (II) ION, Methionine aminopeptidase, ... | | Authors: | Scheidig, A.J, Altmeyer, M, Klein, C.D. | | Deposit date: | 2014-05-23 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Beta-aminoketones as prodrugs for selective irreversible inhibitors of type-1 methionine aminopeptidases.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N96
 
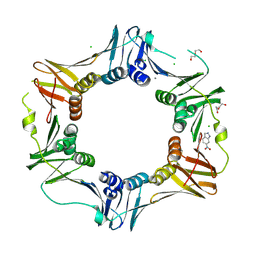 | | E. coli sliding clamp in complex with 6-nitroindazole | | Descriptor: | 6-NITROINDAZOLE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N9A
 
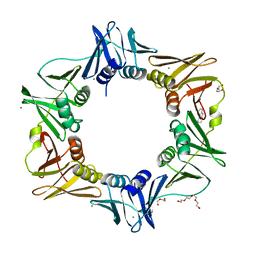 | | E. coli sliding clamp in complex with (R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid | | Descriptor: | (1R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N97
 
 | | E. coli sliding clamp in complex with 5-nitroindole | | Descriptor: | 5-nitro-1H-indole, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
4N95
 
 | | E. coli sliding clamp in complex with 5-chloroindoline-2,3-dione | | Descriptor: | 5-chloro-1H-indole-2,3-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
7DPS
 
 | |
3F1V
 
 | | E. coli Beta Sliding Clamp, 148-153 Ala Mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA polymerase III subunit beta | | Authors: | Cody, V. | | Deposit date: | 2008-10-28 | | Release date: | 2009-09-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Sliding clamp-DNA interactions are required for viability and contribute to DNA polymerase management in Escherichia coli.
J.Mol.Biol., 387, 2009
|
|
3A8C
 
 | |
1YQN
 
 | | E. coli ispF double mutant | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-DIPHOSPHATE, GERANYL DIPHOSPHATE, ... | | Authors: | Sgraja, T, Kemp, L.E, Ramsden, N, Hunter, W.N. | | Deposit date: | 2005-02-02 | | Release date: | 2005-07-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | A double mutation of Escherichia coli2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase disrupts six hydrogen bonds with, yet fails to prevent binding of, an isoprenoid diphosphate.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
4WWL
 
 | | E. coli 5'-nucleotidase mutant I521C labeled with MTSL (intermediate form) | | Descriptor: | CARBONATE ION, GLYCEROL, Protein UshA, ... | | Authors: | Krug, U, Paithankar, K.S, Schultz-Heienbrok, R, Strater, N. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | E. coli 5'-nucleotidase mutant I521C labeled with MTSL (intermediate form)
To Be Published
|
|
1UAA
 
 | | E. COLI REP HELICASE/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), PROTEIN (ATP-DEPENDENT DNA HELICASE REP.) | | Authors: | Korolev, S, Waksman, G. | | Deposit date: | 1997-06-30 | | Release date: | 1998-07-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP.
Cell(Cambridge,Mass.), 90, 1997
|
|
1QFF
 
 | | E. COLI FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) IN COMPLEX WITH BOUND FERRICHROME-IRON | | Descriptor: | 2-AMINO-VINYL-PHOSPHATE, 3-HYDROXY-TETRADECANOIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Ferguson, A.D, Hofmann, E, Coulton, J.W, Diederichs, K, Welte, W. | | Deposit date: | 1999-04-10 | | Release date: | 2000-07-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins.
Structure Fold.Des., 8, 2000
|
|
4XK4
 
 | | E. coli transcriptional regulator RUTR with dihydrouracil | | Descriptor: | DIHYDROPYRIMIDINE-2,4(1H,3H)-DIONE, HTH-type transcriptional regulator RutR | | Authors: | Shumilin, I.A, Cooper, D.R, Shabalin, I.G, Grabowski, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-01-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | E. COLI TRANSCRIPTIONAL REGULATOR RUTR WITH DIHYDROURACIL
to be published
|
|
