5E2M
 
 | | Crystal structure of human carbonic anhydrase isozyme I with 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2015-10-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Intrinsic Thermodynamics and Structures of 2,4- and 3,4-Substituted Fluorinated Benzenesulfonamides Binding to Carbonic Anhydrases.
ChemMedChem, 12, 2017
|
|
4Q30
 
 | | Nitrowillardiine bound to the ligand binding domain of GluA2 at pH 3.5 | | Descriptor: | 3-(5-nitro-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-L-alanine, Glutamate receptor 2 CHIMERIC PROTEIN, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2014-04-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Thermodynamics and mechanism of the interaction of willardiine partial agonists with a glutamate receptor: implications for drug development.
Biochemistry, 53, 2014
|
|
8AHC
 
 | | Crystal structure of the BRD9 bromodomain with BI-7189 | | Descriptor: | Bromodomain-containing protein 9, [2,6-dimethoxy-4-(1,2,5-trimethyl-6-oxidanylidene-pyridin-3-yl)phenyl]methyl-dimethyl-azanium | | Authors: | Bader, G, Boettcher, J, Weiss-Puxbaum, A. | | Deposit date: | 2022-07-21 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Discovery of a Chemical Probe to Study Implications of BPTF Bromodomain Inhibition in Cellular and in vivo Experiments.
Chemmedchem, 18, 2023
|
|
4XZN
 
 | | Crystal structure of the methylated K125R/V301L AKR1B10 Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZL
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0049 | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Dominguez, M, de Lera, A.R, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
4XZM
 
 | | Crystal structure of the methylated wild-type AKR1B10 holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Podjarny, A. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Determinants of the Selectivity of 3-Benzyluracil-1-acetic Acids toward Human Enzymes Aldose Reductase and AKR1B10.
Chemmedchem, 10, 2015
|
|
6VNQ
 
 | |
2ADO
 
 | | Crystal Structure Of The Brct Repeat Region From The Mediator of DNA damage checkpoint protein 1, MDC1 | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Lee, M.S, Edwards, R.A, Thede, G.L, Glover, J.N. | | Deposit date: | 2005-07-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the BRCT Repeat Domain of MDC1 and Its Specificity for the Free COOH-terminal End of the {gamma}-H2AX Histone Tail.
J.Biol.Chem., 280, 2005
|
|
4OYO
 
 | | Human solAC Complexed with 4-(2-Chlorophenyl)-3-methyl-1H-pyrazole | | Descriptor: | 4-(2-chlorophenyl)-5-methyl-1H-pyrazole, Adenylate cyclase type 10, GLYCEROL | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-12 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
4OZ3
 
 | | Human solAC Complexed with 4-phenyl-3-(trifluoromethyl)-1H-pyrazole | | Descriptor: | 4-phenyl-3-(trifluoromethyl)-1H-pyrazole, Adenylate cyclase type 10, GLYCEROL | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
4OYB
 
 | | Crystal Structure Analysis of the solAC | | Descriptor: | Adenylate cyclase type 10, GLYCEROL, ethyl 2-[3-[(4-azanyl-1,2,5-oxadiazol-3-yl)carbonyl]phenoxy]ethanoate | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-11 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
4OYP
 
 | | Human solAC Complexed with 1-Benzofuran-2-carboxylic acid | | Descriptor: | 1-benzofuran-2-carboxylic acid, Adenylate cyclase type 10, CHLORIDE ION | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-12 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
4OZ2
 
 | | Human solAC Complexed with 4-(4-Fluorophenyl)-3-methyl-1H-pyrazole | | Descriptor: | 4-(4-fluorophenyl)-3-methyl-1H-pyrazole, Adenylate cyclase type 10, CHLORIDE ION, ... | | Authors: | Vinkovic, M. | | Deposit date: | 2014-02-13 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human soluble adenylate cyclase reveals a distinct, highly flexible allosteric bicarbonate binding pocket.
Chemmedchem, 9, 2014
|
|
4P90
 
 | | Crystal structure of the kinase domain of human PAK1 in complex with compound 15 | | Descriptor: | Serine/threonine-protein kinase PAK 1, [2-chloro-5-(hydroxymethyl)phenyl]{5-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-1H-pyrrolo[2,3-b]pyridin-3-yl}methanone | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-04-01 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Identification and optimisation of 7-azaindole PAK1 inhibitors with improved potency and kinase selectivity.
MEDCHEMCOMM, 2014
|
|
2ZN7
 
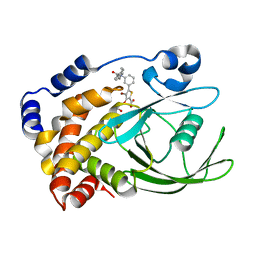 | | CRYSTAL STRUCTURES OF PTP1B-Inhibitor Complexes | | Descriptor: | 4-bromo-3-(carboxymethoxy)-5-{3-[cyclohexyl(phenylcarbonyl)amino]phenyl}thiophene-2-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Wu, J. | | Deposit date: | 2008-04-22 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase-1 B inhibitors: capturing interactions with arginine 24
Chemmedchem, 3, 2008
|
|
2MOF
 
 | | Structural insights of TM domain of LAMP-2A in DPC micelles | | Descriptor: | Lysosome-associated membrane glycoprotein 2 | | Authors: | Tjandra, N, Rout, A. | | Deposit date: | 2014-04-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Transmembrane Domain of Lysosome-associated Membrane Protein Type 2a (LAMP-2A) Reveals Key Features for Substrate Specificity in Chaperone-mediated Autophagy.
J.Biol.Chem., 289, 2014
|
|
2MOM
 
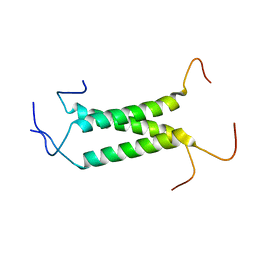 | | Structural insights of TM domain of LAMP-2A in DPC micelles | | Descriptor: | Lysosome-associated membrane glycoprotein 2 | | Authors: | Tjandra, N, Rout, A. | | Deposit date: | 2014-04-27 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Transmembrane Domain of Lysosome-associated Membrane Protein Type 2a (LAMP-2A) Reveals Key Features for Substrate Specificity in Chaperone-mediated Autophagy.
J.Biol.Chem., 289, 2014
|
|
1ZGA
 
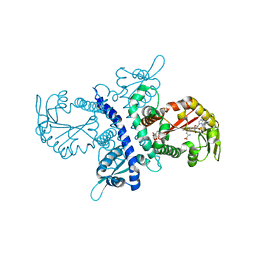 | | Crystal structure of isoflavanone 4'-O-methyltransferase complexed with (+)-6a-hydroxymaackiain | | Descriptor: | (6AR,12AR)-6H-[1,3]DIOXOLO[5,6][1]BENZOFURO[3,2-C]CHROMENE-3,6A(12AH)-DIOL, Isoflavanone 4'-O-methyltransferase', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-20 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
2ZMM
 
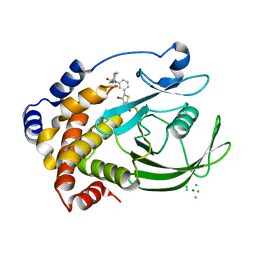 | | Crystal structure of PTP1B-inhibitor complex | | Descriptor: | 4-bromo-3-(carboxymethoxy)-5-{3-[cyclohexyl(methylcarbamoyl)amino]phenyl}thiophene-2-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Xu, W, Wu, J. | | Deposit date: | 2008-04-19 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase-1 B inhibitors: capturing interactions with arginine 24
Chemmedchem, 3, 2008
|
|
5UA4
 
 | |
5UA5
 
 | |
5WOS
 
 | |
4YIH
 
 | | Crystal structure of human cytosolic 5'(3')-deoxyribonucleotidase in complex with the inhibitor PB-PVU | | Descriptor: | 1-{2-deoxy-3,5-O-[phenyl(phosphono)methylidene]-beta-D-threo-pentofuranosyl}-5-[(E)-2-phosphonoethenyl]pyrimidine-2,4(1H,3H)-dione, 5'(3')-deoxyribonucleotidase, cytosolic type, ... | | Authors: | Pachl, P, Rezacova, P, Brynda, J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-based design of a bisphosphonate 5'(3')-deoxyribonucleotidase inhibitor
Medchemcomm, 6, 2015
|
|
5Z9E
 
 | |
5Z9N
 
 | |
