4OSW
 
 | | Crystal structure of the S505E mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3, ... | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
4OTD
 
 | | Crystal Structure of PRK1 Catalytic Domain | | Descriptor: | Serine/threonine-protein kinase N1 | | Authors: | Chamberlain, P.P, Delker, S, Pagarigan, B, Mahmoudi, A, Jackson, P, Abbassian, M, Muir, J, Raheja, N, Cathers, B. | | Deposit date: | 2014-02-13 | | Release date: | 2014-08-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of PRK1 in Complex with the Clinical Compounds Lestaurtinib and Tofacitinib Reveal Ligand Induced Conformational Changes.
Plos One, 9, 2014
|
|
4OX0
 
 | |
1ILU
 
 | | X-RAY CRYSTAL STRUCTURE THE TWO SITE-SPECIFIC MUTANTS ILE7SER AND PHE110SER OF AZURIN FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Hammann, C, Nar, H, Huber, R, Messerschmidt, A. | | Deposit date: | 1995-10-12 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the two site-specific mutants Ile7Ser and Phe110Ser of azurin from Pseudomonas aeruginosa.
J.Mol.Biol., 255, 1996
|
|
1WTC
 
 | | Crystal Structure of S.pombe Serine Racemase complex with AMPPCP | | Descriptor: | Hypothetical protein C320.14 in chromosome III, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Goto, M, Miyahara, I, Hirotsu, K. | | Deposit date: | 2004-11-22 | | Release date: | 2005-11-01 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a homolog of mammalian serine racemase from Schizosaccharomyces pombe
J.Biol.Chem., 284, 2009
|
|
5ER7
 
 | | Connexin-26 Bound to Calcium | | Descriptor: | CALCIUM ION, Gap junction beta-2 protein | | Authors: | Purdy, M.D, Bennett, B.C, Baker, K.A, Yeager, M.J. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.286 Å) | | Cite: | An electrostatic mechanism for Ca(2+)-mediated regulation of gap junction channels.
Nat Commun, 7, 2016
|
|
4ZIR
 
 | | Crystal structure of EcfAA' heterodimer bound to AMPPNP | | Descriptor: | CHLORIDE ION, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Karpowich, N.K, Cocco, N, Song, J.M, Wang, D.N. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ATP binding drives substrate capture in an ECF transporter by a release-and-catch mechanism.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4PRX
 
 | |
2YVC
 
 | |
1WC0
 
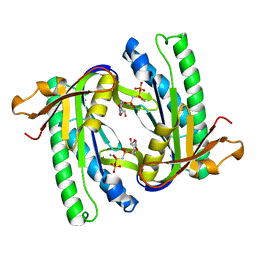 | | Soluble adenylyl cyclase CyaC from S. platensis in complex with alpha, beta-methylene-ATP | | Descriptor: | ADENYLATE CYCLASE, CALCIUM ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Steegborn, C, Litvin, T.N, Levin, L.R, Buck, J, Wu, H. | | Deposit date: | 2004-11-05 | | Release date: | 2004-12-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bicarbonate Activation of Adenylyl Cyclase Via Promotion of Catalytic Active Site Closure and Metal Recruitment
Nat.Struct.Mol.Biol., 12, 2005
|
|
4ZTD
 
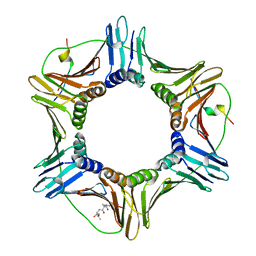 | | Crystal Structure of Human PCNA in complex with a TRAIP peptide | | Descriptor: | ALA-GLY-ALA-GLY-ALA, ALA-PHE-GLN-ALA-LYS-LEU-ASP-THR-PHE-LEU-TRP-SER, Proliferating cell nuclear antigen | | Authors: | Montoya, G, Mortuza, G.B, Blanco, F.J, Ibanez de Opakua, A. | | Deposit date: | 2015-05-14 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress.
J.Cell Biol., 212, 2016
|
|
1ILS
 
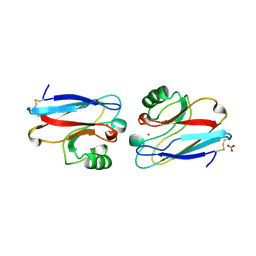 | | X-RAY CRYSTAL STRUCTURE THE TWO SITE-SPECIFIC MUTANTS ILE7SER AND PHE110SER OF AZURIN FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | AZURIN, COPPER (II) ION, NITRATE ION | | Authors: | Hammann, C, Nar, H, Huber, R, Messerschmidt, A. | | Deposit date: | 1995-10-12 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of the two site-specific mutants Ile7Ser and Phe110Ser of azurin from Pseudomonas aeruginosa.
J.Mol.Biol., 255, 1996
|
|
5EPV
 
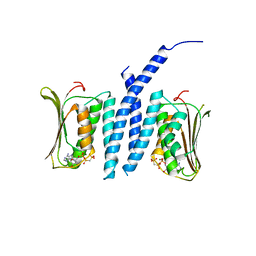 | | Histidine kinase domain from the LOV-HK blue-light receptor from Brucella abortus | | Descriptor: | Blue-light-activated histidine kinase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Rinaldi, J, Guimaraes, B.G, Legrand, P, Thompson, A, Paris, G, Goldbaum, F.A, Klinke, S. | | Deposit date: | 2015-11-12 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Insights into the HWE Histidine Kinase Family: The Brucella Blue Light-Activated Histidine Kinase Domain.
J.Mol.Biol., 428, 2016
|
|
5I1M
 
 | | Yeast V-ATPase average of densities, a subunit segment | | Descriptor: | V-type proton ATPase subunit a, vacuolar isoform | | Authors: | Schep, D.G, Zhao, J, Rubinstein, J.L. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ES3
 
 | | Co-crystal structure of LDH liganded with oxamate | | Descriptor: | L-lactate dehydrogenase A chain, OXAMIC ACID | | Authors: | Nowicki, M.W, Wear, M.A, McNae, I.W, Blackburn, E.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Streamlined, Automated Protocol for the Production of Milligram Quantities of Untagged Recombinant Rat Lactate Dehydrogenase A Using AKTAxpressTM.
Plos One, 10, 2015
|
|
4PU9
 
 | |
1IG9
 
 | | Structure of the Replicating Complex of a Pol Alpha Family DNA Polymerase | | Descriptor: | 5'-D(*AP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*(DOC))-3', CALCIUM ION, ... | | Authors: | Franklin, M.C, Wang, J, Steitz, T.A. | | Deposit date: | 2001-04-17 | | Release date: | 2001-06-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Replicating Complex of a Pol Alpha Family DNA Polymerase
Cell(Cambridge,Mass.), 105, 2001
|
|
3LY2
 
 | | Catalytic Domain of Human Phosphodiesterase 4B in Complex with A Coumarin-Based Inhibitor | | Descriptor: | 8-(cyclopentyloxy)-4-[(3,5-dichloropyridin-4-yl)amino]-7-methoxy-2H-chromen-2-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Shiau, A.K, Coyle, A.R, Hsien, J.H, Staszewski, L.M. | | Deposit date: | 2010-02-26 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Water-soluble PDE4 inhibitors for the treatment of dry eye.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2YAZ
 
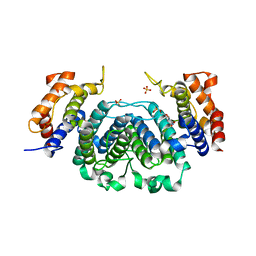 | | The Crystal Structure of Leishmania major dUTPase in complex dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Hemsworth, G.R, Moroz, O.V, Fogg, M.J, Scott, B, Bosch-Navarrete, C, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
4PGM
 
 | |
2YAY
 
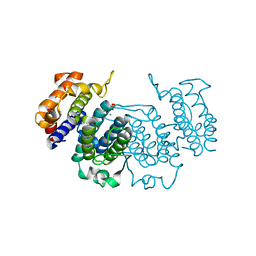 | | The Crystal Structure of Leishmania major dUTPase in complex with substrate analogue dUpNpp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, DUTPASE | | Authors: | Hemsworth, G.R, Moroz, O.V, Fogg, M.J, Scott, B, Bosch-Navarrete, C, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
5ETJ
 
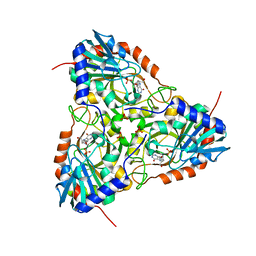 | | Crystal structure of purine nucleoside phosphorylase (E258D, L261A) mutant from human complexed with DADMe-ImmG and phosphate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Suarez, J, Schramm, V.L, Almo, S.C. | | Deposit date: | 2015-11-17 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modulating Enzyme Catalysis through Mutations Designed to Alter Rapid Protein Dynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5EXA
 
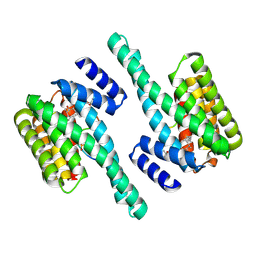 | | Small-molecule stabilization of the 14-3-3/Gab2 PPI interface | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, Fusicoccin A-THF derivative, ... | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2015-11-23 | | Release date: | 2016-05-04 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small-Molecule Stabilization of the 14-3-3/Gab2 Protein-Protein Interaction (PPI) Interface.
Chemmedchem, 11, 2016
|
|
3LUT
 
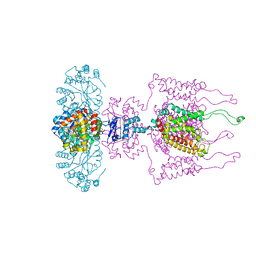 | | A Structural Model for the Full-length Shaker Potassium Channel Kv1.2 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2, ... | | Authors: | Chen, X, Ni, F, Wang, Q, Ma, J. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4PRV
 
 | |
