3MK8
 
 | |
6P94
 
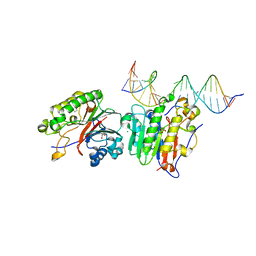 | | Human APE1 C65A AP-endonuclease product complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whitaker, A.W, Stark, W.J, Freudenthal, B.D. | | Deposit date: | 2019-06-09 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Functions of the major abasic endonuclease (APE1) in cell viability and genotoxin resistance.
Mutagenesis, 35, 2020
|
|
7PP1
 
 | |
6P9G
 
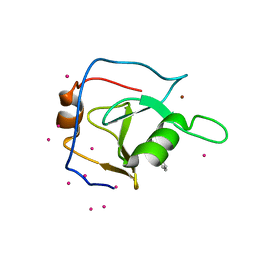 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 2-(4-oxoquinazolin-3(4H)-yl)propanoic acid | | Descriptor: | (2R)-2-(4-oxoquinazolin-3(4H)-yl)propanoic acid, UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 5, ... | | Authors: | Tempel, W, Mann, M.K, Harding, R.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-10 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Small Molecule Antagonists of the USP5 Zinc Finger Ubiquitin-Binding Domain.
J.Med.Chem., 62, 2019
|
|
8RJD
 
 | |
7A3M
 
 | |
6PAU
 
 | |
3ZGE
 
 | | Greater efficiency of photosynthetic carbon fixation due to single amino acid substitution | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, C4 PHOSPHOENOLPYRUVATE CARBOXYLASE, ... | | Authors: | Paulus, J.K, Schlieper, D, Groth, G. | | Deposit date: | 2012-12-17 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Greater Efficiency of Photosynthetic Carbon Fixation due to Single Amino Acid Substitution
Nat.Commun., 4, 2013
|
|
6PAO
 
 | |
7A5K
 
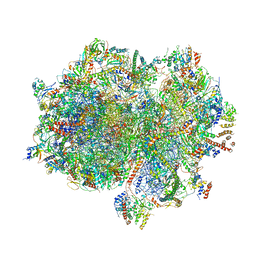 | | Structure of the human mitoribosome in the post translocation state bound to mtEF-G1 | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
8RBX
 
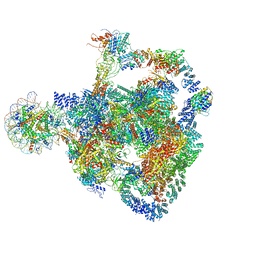 | | Structure of Integrator-PP2A bound to a paused RNA polymerase II-DSIF-NELF-nucleosome complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
3MN7
 
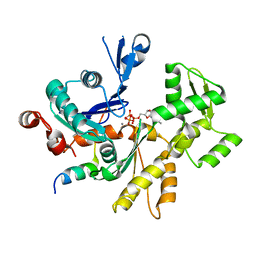 | | Structures of actin-bound WH2 domains of Spire and the implication for filament nucleation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Ducka, A.M, Sitar, T, Popowicz, G.M, Huber, R, Holak, T.A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6PCM
 
 | |
8RJB
 
 | |
5EJ3
 
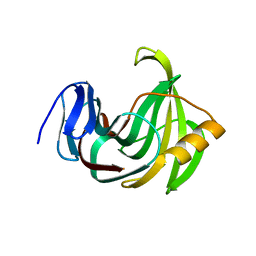 | | Crystal structure of XlnB2 | | Descriptor: | Endo-1,4-beta-xylanase B | | Authors: | Couture, J.-F. | | Deposit date: | 2015-11-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.314 Å) | | Cite: | Ligand Binding Enhances Millisecond Conformational Exchange in Xylanase B2 from Streptomyces lividans.
Biochemistry, 55, 2016
|
|
3ZLN
 
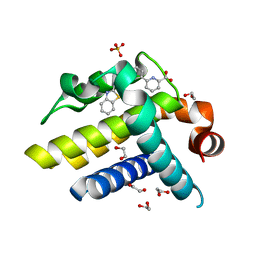 | | Crystal structure of BCL-XL in complex with inhibitor (Compound 3) | | Descriptor: | 1,2-ETHANEDIOL, 6-[(8E)-8-(1,3-benzothiazol-2-ylhydrazinylidene)-6,7-dihydro-5H-naphthalen-2-yl]pyridine-2-carboxylic acid, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
6PD8
 
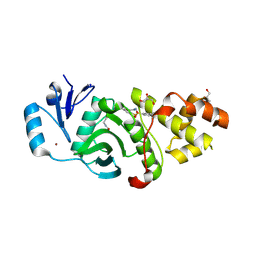 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 39 | | Descriptor: | 5-ethoxy-2-fluoro-3-methyl-N'-(phenylsulfonyl)benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
6PDC
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 42 | | Descriptor: | 2-fluoro-3-methyl-N'-(phenylsulfonyl)-5-[(prop-2-en-1-yl)oxy]benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
5EJX
 
 | | X-ray Free Electron Laser Structure of Cytochrome C Peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PHOSPHATE ION, ... | | Authors: | Doukov, T, Soltis, S.M, Baxter, E.L, Cohen, A, Song, J, McPhillips, S, Poulos, T.L, Meharenna, Y.T, Chreifi, G. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the pristine peroxidase ferryl center and its relevance to proton-coupled electron transfer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7AEA
 
 | | Cryo-EM structure of human RNA Polymerase III elongation complex 2 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7A97
 
 | | SARS-CoV-2 Spike Glycoprotein with 2 ACE2 Bound | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
5EKX
 
 | | DENGUE 3 NS5 METHYLTRANSFERASE BOUND TO S-ADENOSYLMETHIONINE AND FRAGMENT NB2E11 | | Descriptor: | 4-chloro-5-methylbenzene-1,2-diamine, NS5 METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Barral, K, Bricogne, G, Sharff, A. | | Deposit date: | 2015-11-04 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of novel dengue virus NS5 methyltransferase non-nucleoside inhibitors by fragment-based drug design.
Eur.J.Med.Chem., 125, 2016
|
|
5GW6
 
 | |
5GWF
 
 | | FraC with GlcNAc(6S) bound | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, CHLORIDE ION, DELTA-actitoxin-Afr1a, ... | | Authors: | Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2016-09-11 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Haemolytic actinoporins interact with carbohydrates using their lipid-binding module
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 372, 2017
|
|
6PGK
 
 | | Membrane Protein Megahertz Crystallography at the European XFEL, Photosystem I XFEL at 2.9 A | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Gisriel, C, Fromme, P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Membrane protein megahertz crystallography at the European XFEL.
Nat Commun, 10, 2019
|
|
