9Q8L
 
 | | Crystal Structure of 21A08Ap1-Fab in Complex with Human PD-1 at 1.85 angstrom Resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Murer, P, Petersen, L, Egli, N, Salazar, U, Neubert, P, Zurbach, A, Rau, A, Stocker, C, Katopodis, A, Huber, C, Carr, K, Arduin, A. | | Deposit date: | 2025-02-25 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | ANV600 is a novel PD-1 targeted IL-2R beta gamma agonist that selectively expands tumor antigen-specific T cells and potentiates PD-1 checkpoint inhibitor therapy.
J Immunother Cancer, 13, 2025
|
|
6OXR
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR-82 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[({4-[(1S)-1-hydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXU
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR-99 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{({4-[(1R)-1-hydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OY2
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-25 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-ethylbutyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
8FUI
 
 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
6PJN
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-41 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3S,5S)-3-hydroxy-5-({1-[(methoxycarbonyl)amino]cyclopentane-1-carbonyl}amino)-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6PJE
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-43 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-({1-[(methoxycarbonyl)amino]cyclopentane-1-carbonyl}amino)-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6WBM
 
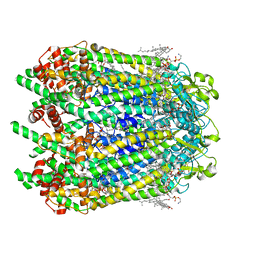 | | Cryo-EM structure of human Pannexin 1 channel N255A mutant | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, DIACYL GLYCEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
6WBF
 
 | | Cryo-EM structure of wild type human Pannexin 1 channel | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
7OUF
 
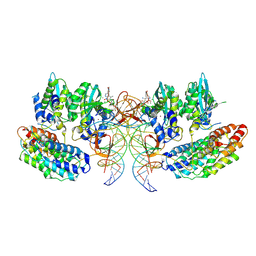 | | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor XZ450 | | Descriptor: | 4-azanyl-~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-6-[3-(dimethylamino)-3-oxidanylidene-propyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7OLX
 
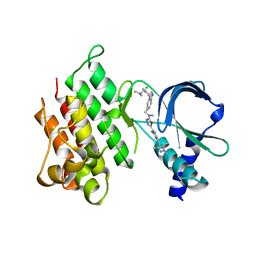 | | MerTK kinase domain with type 1.5 inhibitor containing a tri-methyl pyrazole group | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer, ~{N}-[[3-[4-[(dimethylamino)methyl]phenyl]imidazo[1,2-a]pyridin-6-yl]methyl]-~{N}-methyl-5-[3-methyl-5-(1,3,5-trimethylpyrazol-4-yl)pyridin-2-yl]-1,3,4-oxadiazol-2-amine | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
6O3Z
 
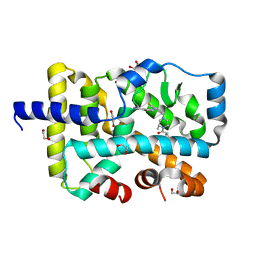 | | Crystal structure of RORgt with 3-cyano-N-(3-{[(3S)-4-(cyclopentanecarbonyl)-3-methylpiperazin-1-yl]methyl}-5-fluoro-2-methylphenyl)benzamide (compound 1) | | Descriptor: | 1,2-ETHANEDIOL, 3-cyano-N-(3-{[(3S)-4-(cyclopentanecarbonyl)-3-methylpiperazin-1-yl]methyl}-5-fluoro-2-methylphenyl)benzamide, RAR-related orphan receptor C isoform a variant | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of [1,2,4]Triazolo[1,5-a]pyridine Derivatives as Potent and Orally Bioavailable ROR gamma t Inverse Agonists.
Acs Med.Chem.Lett., 11, 2020
|
|
8GMD
 
 | | CRYSTAL STRUCTURE OF AP2 ASSOCIATED KINASE 1 COMPLEXED WITH (5P)-3-({(8R)-5-[(4-aminopiperidin-1-yl)methyl]pyrrolo[2,1-f][1,2,4]triazin-4-yl}amino)-5-[2-(propan-2-yl)-2H-tetrazol-5-yl]phenol | | Descriptor: | (5P)-3-({(8R)-5-[(4-aminopiperidin-1-yl)methyl]pyrrolo[2,1-f][1,2,4]triazin-4-yl}amino)-5-[2-(propan-2-yl)-2H-tetrazol-5-yl]phenol, AP2-associated protein kinase 1, SULFATE ION | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of pyrrolo[2,1- f ][1,2,4]triazine-based inhibitors of adaptor protein 2-associated kinase 1 for the treatment of pain.
Med.Chem.Res., 2023
|
|
6OXO
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-91 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[{[4-(hydroxymethyl)phenyl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXP
 
 | | HIV-1 Protease NL4-3 WT in Complex with UMass3 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)propyl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OTG
 
 | | HIV-1 protease triple mutants V32I, I47V, V82I with GRL-011-11A (a methylamine bis-Tetrahydrofuran P2-Ligand, sulfonamide isostere derivate) | | Descriptor: | (3R,3aS,4R,6aR)-4-(methylamino)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Pawar, S, Weber, I.T. | | Deposit date: | 2019-05-03 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural studies of antiviral inhibitor with HIV-1 protease bearing drug resistant substitutions of V32I, I47V and V82I.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
6OOS
 
 | | HIV-1 Protease NL4-3 L90M Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, NL4-3 PROTEASE, SULFATE ION | | Authors: | Henes, M, Kosovrasti, K, Lockbaum, G.J, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A, Whitfield, T.W. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Determinants of Epistasis in HIV-1 Protease: Elucidating the Interdependence of L89V and L90M Mutations in Resistance.
Biochemistry, 58, 2019
|
|
6ZFH
 
 | | Structure of human galactokinase in complex with galactose and 2'-(benzo[d]oxazol-2-ylamino)-7',8'-dihydro-1'H-spiro[cyclopentane-1,4'-quinazolin]-5'(6'H)-one | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclopentane]-5-one, Galactokinase, beta-D-galactopyranose | | Authors: | Bezerra, G.A, Mackinnon, S, Zhang, M, Foster, W, Bailey, H, Arrowsmith, C, Edwards, A, Bountra, C, Lai, K, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.439 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
6OXQ
 
 | | HIV-1 Protease NL4-3 WT in Complex with UMass8 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-1-benzyl-3-[(2-ethylbutyl){[4-(hydroxymethyl)phenyl]sulfonyl}amino]-2-hydroxypropyl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6WGD
 
 | | Crystal structure of a 6-phospho-beta-glucosidase from Bacillus licheniformis | | Descriptor: | 1,2-ETHANEDIOL, 6-phospho-beta-glucosidase | | Authors: | Liberato, M.V, Popov, A, Polikarpov, I. | | Deposit date: | 2020-04-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Structure, Bioinformatics Analysis, and Substrate Specificity of a 6-Phospho-beta-glucosidase Glycoside Hydrolase 1 Enzyme from Bacillus licheniformis .
J.Chem.Inf.Model., 60, 2020
|
|
6XNQ
 
 | |
6PJI
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR3-43 | | Descriptor: | Protease NL4-3, SULFATE ION, methyl [(1S)-1-cyclopropyl-2-({(2S,3S,5S)-5-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-1,6-diphenylhexan-2-yl}amino)-2-oxoethyl]carbamate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
9RUB
 
 | | CRYSTAL STRUCTURE OF ACTIVATED RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE COMPLEXED WITH ITS SUBSTRATE, RIBULOSE-1,5-BISPHOSPHATE | | Descriptor: | FORMIC ACID, MAGNESIUM ION, RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE, ... | | Authors: | Lundqvist, T, Schneider, G. | | Deposit date: | 1990-11-28 | | Release date: | 1993-01-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of activated ribulose-1,5-bisphosphate carboxylase complexed with its substrate, ribulose-1,5-bisphosphate.
J.Biol.Chem., 266, 1991
|
|
7Q4I
 
 | | Crystal structure of DmC1GalT1 in complex with UDP-Mn2+ and the APD-TGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, Glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase 1, ... | | Authors: | Gonzalez-Ramirez, A.M, Coelho, H, Companon, I, Grosso, A.S, Yang, Z, Narimatsu, Y, Clausen, H, Marcelo, F, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2021-10-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the synthesis of the core 1 structure by C1GalT1.
Nat Commun, 13, 2022
|
|
9QB5
 
 | | AP2-associated protein kinase 1 (AAK1) bound to CKJB68 | | Descriptor: | AP2-associated protein kinase 1, SULFATE ION, ~{N}-(phenylmethyl)-7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxamide | | Authors: | Preuss, F, Mensing, T.E, Hanke, T, Knapp, S, Mathea, S. | | Deposit date: | 2025-02-28 | | Release date: | 2025-04-09 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of pyrazolo[1,5-a]pyrimidine based macrocyclic kinase inhibitors targeting AAK1.
Eur.J.Med.Chem., 299, 2025
|
|
