4OSV
 
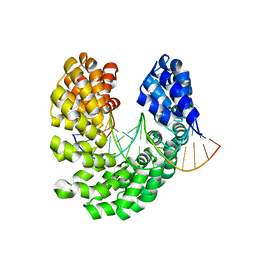 | | Crystal structure of the S505M mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
5NYU
 
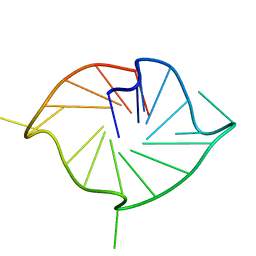 | | M2 G-quadruplex 10 wt% PEG8000 | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
5NYT
 
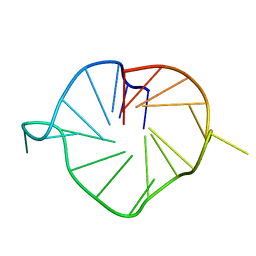 | | M2 G-quadruplex 20 wt% ethylene glycol | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
4OSW
 
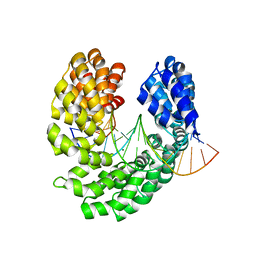 | | Crystal structure of the S505E mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3, ... | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
5WE2
 
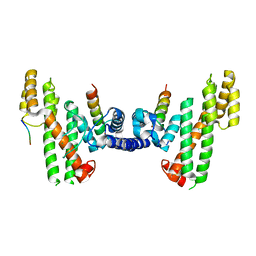 | | Structural Basis for Telomere Length Regulation by the Shelterin Bridge | | Descriptor: | DNA-binding protein rap1, Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ... | | Authors: | Kim, J.-K, Liu, J, Hu, X, Sankaran, B, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
7LV9
 
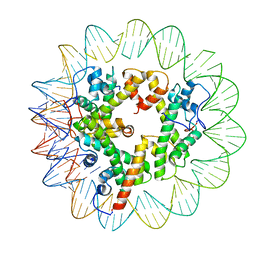 | | Marseillevirus heterotrimeric (hexameric) nucleosome | | Descriptor: | DNA (96-MER), Histone doublet Beta-Alpha (Alpha), Histone doublet Beta-Alpha (Beta), ... | | Authors: | Valencia-Sanchez, M.I, Abini-Agbomson, S, Armache, K.-J. | | Deposit date: | 2021-02-24 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The structure of a virus-encoded nucleosome.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3WVG
 
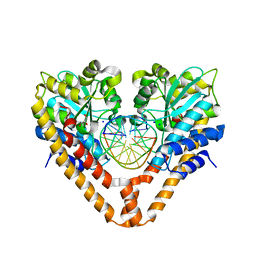 | | Time-Resolved Crystal Structure of HindIII with 0sec soaking | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*GP*CP*TP*TP*GP*GP*C)-3'), GLYCEROL, SODIUM ION, ... | | Authors: | Kawamura, T, Kobayashi, T, Watanabe, N. | | Deposit date: | 2014-05-21 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analysis of the HindIII-catalyzed reaction by time-resolved crystallography
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1E0K
 
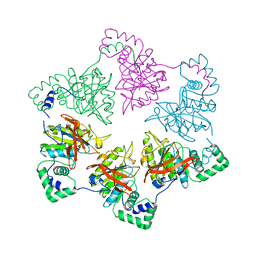 | | gp4d helicase from phage T7 | | Descriptor: | DNA HELICASE | | Authors: | Singleton, M.R, Sawaya, M.R, Ellenberger, T, Wigley, D.B. | | Deposit date: | 2000-03-30 | | Release date: | 2000-06-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of T7 Gene 4 Ring Helicase Indicates a Mechanism for Sequential Hydrolysis of Nucleotides
Cell(Cambridge,Mass.), 101, 2000
|
|
9DP1
 
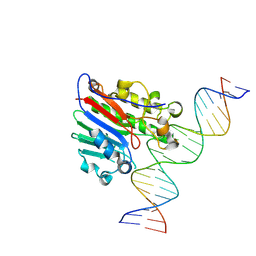 | | APE1 N174A Substrate Complex with Abasic DNA | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*(3DR)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA repair nuclease/redox regulator APEX1, ... | | Authors: | DeHart, K.M, Freudenthal, B.D. | | Deposit date: | 2024-09-20 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | N174 Physically and Chemically Stabilizes the Endonuclease Reaction of APE1
To Be Published
|
|
6L92
 
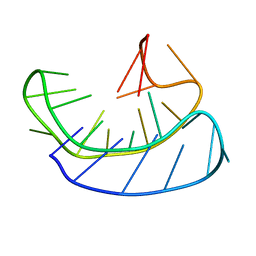 | | A basket type G-quadruplex in WNT DNA promoter | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*CP*AP*CP*CP*GP*GP*GP*CP*AP*GP*TP*GP*GP*GP*CP*GP*GP*G)-3') | | Authors: | Wang, Z.F, Li, M.H, Chu, I.T, Winnerdy, F.R, Phan, A.T, Chang, T.C. | | Deposit date: | 2019-11-08 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cytosine epigenetic modification modulates the formation of an unprecedented G4 structure in the WNT1 promoter.
Nucleic Acids Res., 48, 2020
|
|
6L8M
 
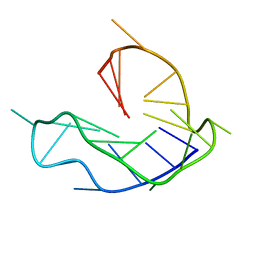 | | WNT DNA promoter mutant G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*CP*AP*CP*CP*GP*GP*GP*CP*AP*GP*TP*GP*GP*GP*CP*GP*GP*G)-3') | | Authors: | Wang, Z.F, Li, M.H, Chu, I.T, Winnerdy, F.R, Phan, A.T, Chang, T.C. | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cytosine epigenetic modification modulates the formation of an unprecedented G4 structure in the WNT1 promoter.
Nucleic Acids Res., 48, 2020
|
|
6RLS
 
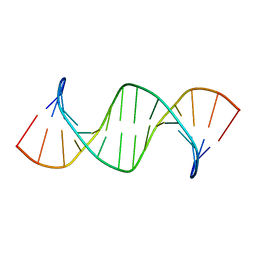 | | Concerted dynamics of metallo-base pairs in an A/B-form helical transition (apo species) | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*TP*CP*AP*TP*GP*AP*TP*AP*CP*G)-3')_apo | | Authors: | Schmidt, O.P, Jurt, S, Johannsen, S, Karimi, A, Sigel, R.K.O, Luedtke, N.W. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Concerted dynamics of metallo-base pairs in an A/B-form helical transition.
Nat Commun, 10, 2019
|
|
4HUE
 
 | | Structure of 5-chlorouracil modified G:U base pair | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(UCL)P*GP*CP*G)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs.
Nucleic Acids Res., 41, 2013
|
|
6AR5
 
 | |
7EJE
 
 | | human RAD51 post-synaptic complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | Authors: | Zhao, L.Y, Xu, J.F, Wang, H.W. | | Deposit date: | 2021-04-02 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Nucleic Acids Res., 49, 2021
|
|
8T9F
 
 | |
1JG7
 
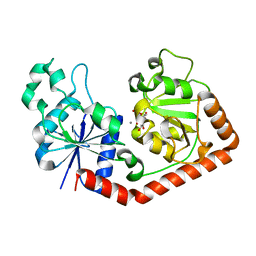 | | T4 phage BGT in complex with UDP and Mn2+ | | Descriptor: | DNA BETA-GLUCOSYLTRANSFERASE, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Lariviere, L, Kurzeck, J, Aschke-Sonnenborn, U, Freemont, P.S, Janin, J, Ruger, W. | | Deposit date: | 2001-06-23 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding.
J.Mol.Biol., 311, 2001
|
|
6FCV
 
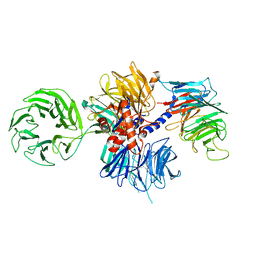 | | Structure of the human DDB1-CSA complex | | Descriptor: | DNA damage-binding protein 1, DNA excision repair protein ERCC-8 | | Authors: | Meulenbroek, E.M, Pannu, N.S. | | Deposit date: | 2017-12-21 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | TRiC controls transcription resumption after UV damage by regulating Cockayne syndrome protein A.
Nat Commun, 9, 2018
|
|
8WWO
 
 | |
1JEJ
 
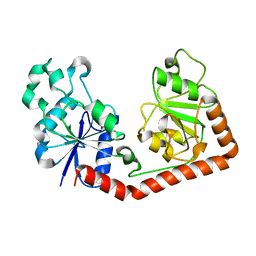 | | T4 phage apo BGT | | Descriptor: | DNA BETA-GLUCOSYLTRANSFERASE | | Authors: | Morera, S, Lariviere, L, Kurzeck, J, Aschke-Sonnenborn, U, Freemont, P.S, Janin, J, Ruger, W. | | Deposit date: | 2001-06-18 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding.
J.Mol.Biol., 311, 2001
|
|
1JIU
 
 | | T4 Phage BGT in Complex with Mg2+ : Form I | | Descriptor: | DNA BETA-GLUCOSYLTRANSFERASE, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Lariviere, L, Kurzeck, J, Aschke-Sonnenborn, U, Freemont, P.S, Janin, J, Ruger, W. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding.
J.Mol.Biol., 311, 2001
|
|
7WLR
 
 | | Cryo-EM structure of the nucleosome containing Komagataella pastoris histones | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B, ... | | Authors: | Fukushima, Y, Hatazawa, S, Hirai, S, Kujirai, T, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-01-13 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and biochemical analyses of the nucleosome containing Komagataella pastoris histones.
J.Biochem., 172, 2022
|
|
1JG6
 
 | | T4 phage BGT in complex with UDP | | Descriptor: | DNA BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Lariviere, L, Kurzeck, J, Aschke-Sonnenborn, U, Freemont, P.S, Janin, J, Ruger, W. | | Deposit date: | 2001-06-23 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding.
J.Mol.Biol., 311, 2001
|
|
1JIV
 
 | | T4 phage BGT in complex with Mg2+ : Form II | | Descriptor: | DNA BETA-GLUCOSYLTRANSFERASE, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Lariviere, L, Kurzeck, J, Aschke-Sonnenborn, U, Freemont, P.S, Janin, J, Ruger, W. | | Deposit date: | 2001-07-03 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding.
J.Mol.Biol., 311, 2001
|
|
8YGH
 
 | | pP1192R-apo open state | | Descriptor: | DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 2024
|
|
