3EEQ
 
 | | Crystal structure of a putative cobalamin biosynthesis protein G homolog from Sulfolobus solfataricus | | Descriptor: | SULFATE ION, putative Cobalamin biosynthesis protein G homolog | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-05 | | Release date: | 2008-09-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative cobalamin biosynthesis protein G homolog from Sulfolobus solfataricus
To be Published
|
|
3EB6
 
 | | Structure of the cIAP2 RING domain bound to UbcH5b | | Descriptor: | Baculoviral IAP repeat-containing protein 3, Ubiquitin-conjugating enzyme E2 D2, ZINC ION | | Authors: | Mace, P.D, Linke, K, Schumacher, F.-R, Smith, C.A, Day, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of the cIAP2 RING Domain Reveal Conformational Changes Associated with Ubiquitin-conjugating Enzyme (E2) Recruitment.
J.Biol.Chem., 283, 2008
|
|
3EBD
 
 | | Structure of inhibited murine iNOS oxygenase domain | | Descriptor: | (2S)-2-methyl-2,3-dihydrothieno[2,3-f][1,4]oxazepin-5-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-27 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3EBW
 
 | | Crystal structure of major allergens, Per a 4 from cockroaches | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Per a 4 allergen | | Authors: | Tan, Y.W, Chan, S.L, Chew, F.T, Sivaraman, J, Mok, Y.K. | | Deposit date: | 2008-08-28 | | Release date: | 2008-12-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of two major allergens, Bla g 4 and Per a 4, from cockroaches and their IgE binding epitopes.
J.Biol.Chem., 284, 2008
|
|
3EG2
 
 | | Crystal structure of the N114Q mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
3EDJ
 
 | | Structural base for cyclodextrin hydrolysis | | Descriptor: | CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Cyclomaltodextrinase, ... | | Authors: | Buedenbender, S, Schulz, G.E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural base for enzymatic cyclodextrin hydrolysis
J.Mol.Biol., 385, 2009
|
|
3EE1
 
 | |
3EF1
 
 | |
3EG1
 
 | |
3EGU
 
 | | Crystal structure of the N114A mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1, SULFATE ION | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
3EHI
 
 | | Crystal Structure of Human Thymidyalte Synthase M190K with Loop 181-197 stabilized in the inactive conformation | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Thymidylate synthase | | Authors: | Lovelace, L.L, Gibson, L.M, Johnson, S.R, Berger, S.H, Lebioda, L. | | Deposit date: | 2008-09-12 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Variants of human thymidylate synthase with loop 181-197 stabilized in the inactive conformation.
Protein Sci., 18, 2009
|
|
3EHZ
 
 | |
3EJI
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36K at cryogenic temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, Thermonuclease | | Authors: | Khangulov, V.S, Schlessman, J.L, Garcia-Moreno, E.B, Benning, M, Isom, D. | | Deposit date: | 2008-09-18 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Consequences of the Ionization of Lysines at 25 Internal Positions in a Protein
To be Published
|
|
3EJO
 
 | |
3EL0
 
 | |
3B2I
 
 | | Iodide derivative of human LFABP | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3B0J
 
 | | M175E mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B2J
 
 | | Iodide derivative of human LFABP | | Descriptor: | Fatty acid-binding protein, liver, IODIDE ION, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Utility of anion and cation combinations for phasing of protein structures.
J.Struct.Funct.Genom., 13, 2012
|
|
3EKP
 
 | | Crystal Structure of the inhibitor Amprenavir (APV) in complex with a multi-drug resistant HIV-1 protease variant (L10I/G48V/I54V/V64I/V82A)Refer: FLAP+ in citation | | Descriptor: | ACETATE ION, PHOSPHATE ION, Protease, ... | | Authors: | Prabu-Jayabalan, M, King, N.M, Bandaranayake, R.M. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
3ELO
 
 | | Crystal Structure of Human Pancreatic Prophospholipase A2 | | Descriptor: | Phospholipase A2, SULFATE ION | | Authors: | Xu, W, Yi, L, Feng, Y, Chen, L, Liu, J. | | Deposit date: | 2008-09-22 | | Release date: | 2009-04-14 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into the activation mechanism of human pancreatic prophospholipase A2
J.Biol.Chem., 284, 2009
|
|
3EM3
 
 | |
3EMK
 
 | |
3EMS
 
 | | Effect of Ariginine on lysozyme | | Descriptor: | ARGININE, CHLORIDE ION, Lysozyme C, ... | | Authors: | Ito, L, Shiraki, K, Matsuura, T, Yamaguhi, H. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Effect of Ariginine on lysozyme
to be published
|
|
3EN1
 
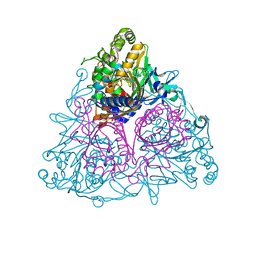 | | Crystal structure of Toluene 2,3-Dioxygenase | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Benzene 1,2-dioxygenase subunit alpha, Benzene 1,2-dioxygenase subunit beta, ... | | Authors: | Friemann, R, Lee, K, Brown, E.N, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2008-09-25 | | Release date: | 2009-03-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the multicomponent Rieske non-heme iron toluene 2,3-dioxygenase enzyme system
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3EP5
 
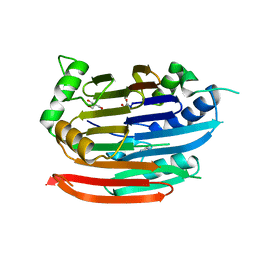 | | Human AdoMetDC E178Q mutant with no putrescine bound | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRUVIC ACID, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
