5OI5
 
 | |
5OI3
 
 | |
5OIA
 
 | |
5OI2
 
 | |
2PSG
 
 | |
1QJZ
 
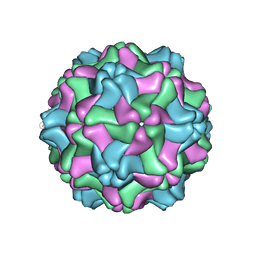 | | Three Dimensional Structure of Physalis Mottle Virus : Implications for the Viral Assembly | | Descriptor: | COAT PROTEIN | | Authors: | Krishna, S.S, Hiremath, C.N, Munshi, S.K, Prahadeeswaran, D, Sastri, M, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 1999-07-07 | | Release date: | 1999-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Three Dimensional Structure of Physalis Movirus: Implications for the Viral Assembly
J.Mol.Biol., 289, 1999
|
|
8RND
 
 | | Cathepsin S in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin S, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2024-01-09 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 2024
|
|
8SXM
 
 | |
8TDO
 
 | |
8TDN
 
 | |
5E5K
 
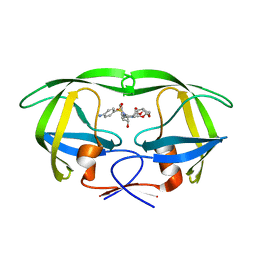 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 4.3 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | Authors: | Kovalevsky, A.Y, Das, A. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.75 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8Q8S
 
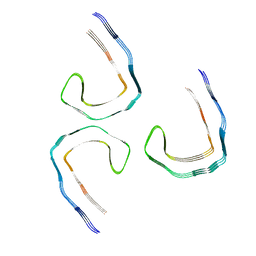 | | Tau - AD-THF | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Li, D, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-18 | | Release date: | 2023-09-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
8Q8Z
 
 | |
8Q9A
 
 | |
8Q8E
 
 | |
8Q8X
 
 | |
8Q9B
 
 | |
8QJJ
 
 | | CTE type II (tau intermediate amyloid) | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Lovestam, S, Scheres, S.H.W. | | Deposit date: | 2023-09-13 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Disease-specific tau filaments assemble via polymorphic intermediates.
Nature, 625, 2024
|
|
5FHL
 
 | |
5HIB
 
 | | EGFR kinase domain mutant "TMLR" with a pyrazolopyrimidine inhibitor | | Descriptor: | Epidermal growth factor receptor, N-tert-butyl-5-{[(1-methyl-1H-pyrazol-5-yl)sulfonyl]amino}pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
5HIC
 
 | | EGFR kinase domain mutant "TMLR" with a imidazopyridinyl-aminopyrimidine inhibitor | | Descriptor: | Epidermal growth factor receptor, N-{2-[1-(cyclopropylsulfonyl)-1H-pyrazol-4-yl]pyrimidin-4-yl}-1-(propan-2-yl)-1H-imidazo[4,5-c]pyridin-6-amine, SULFATE ION | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2.
Cancer Cell, 29, 2016
|
|
5HCH
 
 | | X-ray structure of a lectin-bound DNA duplex containing an unnatural phenanthrenyl pair | | Descriptor: | (6S)-2,6-anhydro-1-deoxy-6-(2-{[(S)-hydroxy(oxido)-lambda~5~-phosphanyl]oxy}ethyl)-D-galactitol, CALCIUM ION, DNA (5'-D(*CP*GP*CP*AP*TP*TP*(DF)P*TP*AP*TP*CP*GP*C)-3'), ... | | Authors: | Roethlisberger, P, Istrate, A, Marcaida Lopez, M.J, Visini, R, Stocker, A, Leumann, C.J. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structure of a lectin-bound DNA duplex containing an unnatural phenanthrenyl pair.
Chem.Commun.(Camb.), 52, 2016
|
|
5FHJ
 
 | |
8AH7
 
 | |
8E7E
 
 | |
