8TD4
 
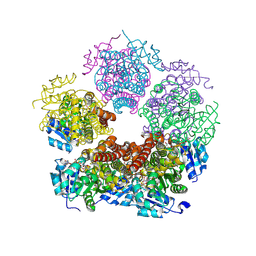 | | Structure of PYCR1 complexed with NADH and 1,3-Dithiolane-2-carboxylic acid | | Descriptor: | 1,3-dithiolane-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
5FXE
 
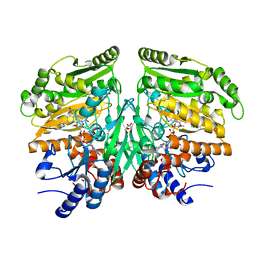 | | Crystal structure of eugenol oxidase in complex with coniferyl alcohol | | Descriptor: | (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enal, EUGENOL OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nguyen, Q.-T, de Gonzalo, G, Binda, C, Martinez, A.R, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2016-03-01 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biocatalytic Properties and Structural Analysis of Eugenol Oxidase from Rhodococcus Jostii Rha1: A Versatile Oxidative Biocatalyst.
Chembiochem, 17, 2016
|
|
5C5E
 
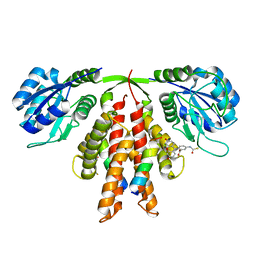 | | Structure of KaiA dimer in complex with C-terminal KaiC peptide at 2.8 A resolution | | Descriptor: | 2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)-5-[(sulfanylcarbonyl)amino]benzoic acid, Circadian clock protein KaiA, KaiC C-terminal peptide | | Authors: | Pattanayek, R, Egli, M. | | Deposit date: | 2015-06-19 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Protein-Protein Interactions in the Cyanobacterial Circadian Clock: Structure of KaiA Dimer in Complex with C-Terminal KaiC Peptides at 2.8 angstrom Resolution.
Biochemistry, 54, 2015
|
|
8TD2
 
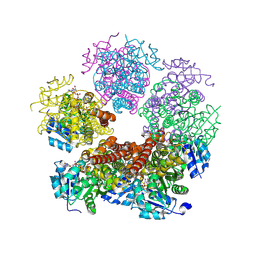 | | Structure of PYCR1 complexed with NADH and cyclobutane-1,1-dicarboxylic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
7TRT
 
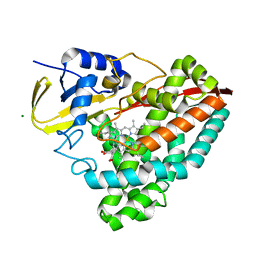 | |
6M7H
 
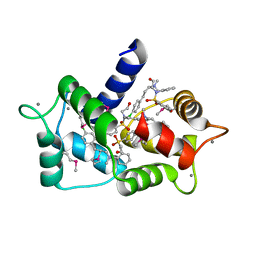 | | Structure of calmodulin with KN93 | | Descriptor: | CALCIUM ION, Calmodulin-1, N-[2-[[[3-(4'-Chlorophenyl)-2-propenyl]methylamino]methyl]phenyl]-N-(2-hydroxyethyl)-4'-methoxybenzenesulfonamide | | Authors: | Damo, S.M, Pattanayek, R, Johnson, C.N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The CaMKII inhibitor KN93-calmodulin interaction and implications for calmodulin tuning of NaV1.5 and RyR2 function.
Cell Calcium, 82, 2019
|
|
8TD7
 
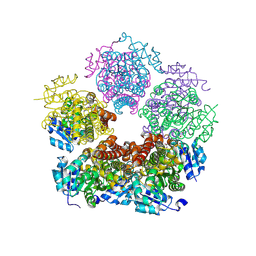 | | Structure of PYCR1 complexed with 2S-hydroxy-3-methylbutyric acid | | Descriptor: | (2S)-2-hydroxy-3-methylbutanoic acid, DI(HYDROXYETHYL)ETHER, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
7TRU
 
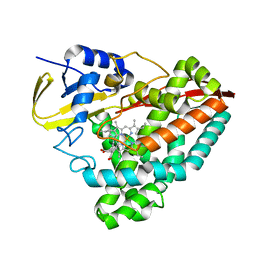 | |
8QGW
 
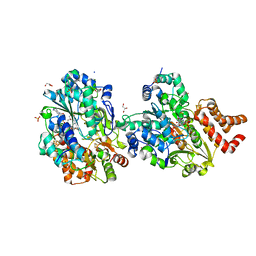 | |
8TDD
 
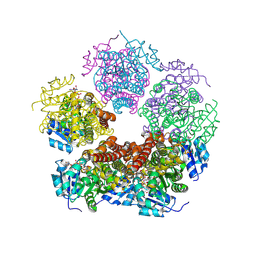 | | Structure of PYCR1 complexed with NADH and 2-(furan-2-yl)acetic acid | | Descriptor: | (furan-2-yl)acetic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
8JQ5
 
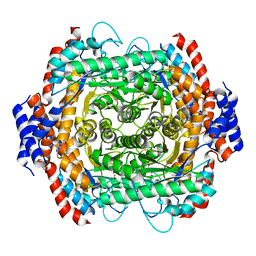 | |
7Y62
 
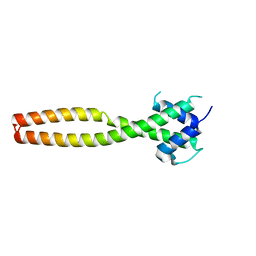 | | Crystal structure of human TFEB HLHLZ domain | | Descriptor: | Transcription factor EB | | Authors: | Yang, G, Li, P, Lin, Y, Liu, Z, Sun, H, Zhao, Z, Fang, P, Wang, J. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A small-molecule drug inhibits autophagy gene expression through the central regulator TFEB.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6M8M
 
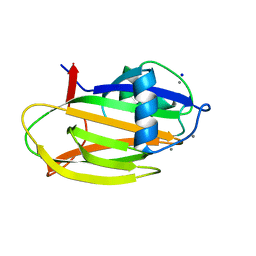 | | PA14 sugar-binding domain from RTX adhesin | | Descriptor: | CALCIUM ION, Putative large adhesion protein (Lap) involved in biofilm formation, SODIUM ION, ... | | Authors: | Vance, T.D.R, Conroy, B, Davies, P.L. | | Deposit date: | 2018-08-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure and functional analysis of a bacterial adhesin sugar-binding domain.
Plos One, 14, 2019
|
|
7OZD
 
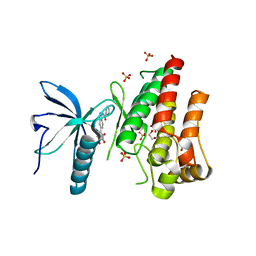 | | FGFR1 kinase domain (residues 458-765) with mutations C488A, C584S in complex with 34. | | Descriptor: | 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, N-[6-(4-hydroxyphenyl)-1H-indazol-3-yl]benzamide, ... | | Authors: | Trinh, C.H, Turner, L.D, Fishwick, C.W.G. | | Deposit date: | 2021-06-27 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | From Fragment to Lead: De Novo Design and Development toward a Selective FGFR2 Inhibitor.
J.Med.Chem., 65, 2022
|
|
5C7W
 
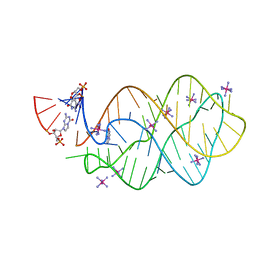 | | 5'-monophosphate Z:P Guanine Riboswitch bound to hypoxanthine. | | Descriptor: | 5'-monophosphate Z:P guanine riboswitch, COBALT HEXAMMINE(III), HYPOXANTHINE | | Authors: | Hernandez, A.R, Shao, Y, Hoshika, S, Yang, Z, Shelke, S.A, Herrou, J, Kim, H.-J, Kim, M.-J, Piccirilli, J.A, Benner, S.A. | | Deposit date: | 2015-06-25 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A Crystal Structure of a Functional RNA Molecule Containing an Artificial Nucleobase Pair.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7U6H
 
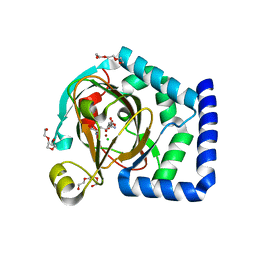 | | HalD with ornithine and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Swenson, C.V, Neugebauer, M.E, Kissman, E.N, Chang, M.C.Y. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biocatalytic control of site-selectivity and chain length-selectivity in radical amino acid halogenases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5TDX
 
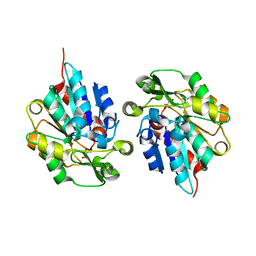 | | Resurrected Ancestral Hydroxynitrile Lyase from Flowering Plants | | Descriptor: | Ancestral Hydroxynitrile Lyase 1, GLYCEROL | | Authors: | Jones, B.J, Evans, R, Wilmot, C.M, Kazlauskas, R.J. | | Deposit date: | 2016-09-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Larger active site in an ancestral hydroxynitrile lyase increases catalytically promiscuous esterase activity.
Plos One, 15, 2020
|
|
8TGF
 
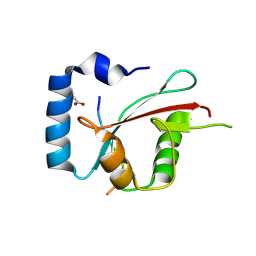 | | Crystal structure of cEPG5 LIR/LGG-1 complex | | Descriptor: | ACETATE ION, Ectopic P granules protein 5, Protein lgg-1 | | Authors: | Cheung, Y.W.S, Yip, C.K. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of the mode of interaction between the tandem LC3-interacting region motif of the autophagy tethering factor EPG5 and ATG8 proteins
To be published
|
|
7YGY
 
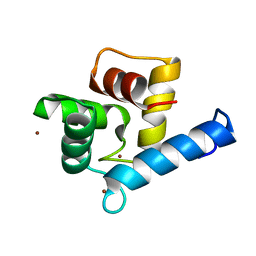 | | Crystal structure of the Zn2+-bound EFhd2/Swiprosin-1 | | Descriptor: | EF-hand domain-containing protein D2, ZINC ION | | Authors: | Mun, S.A, Park, J, Kang, J.Y, Park, T, Jin, M, Yang, J, Eom, S.H. | | Deposit date: | 2022-07-12 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical insights into Zn 2+ -bound EF-hand proteins, EFhd1 and EFhd2.
Iucrj, 10, 2023
|
|
8K2J
 
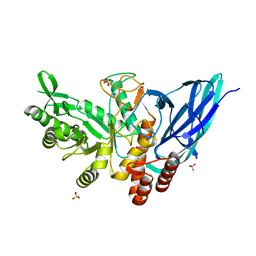 | |
7TZW
 
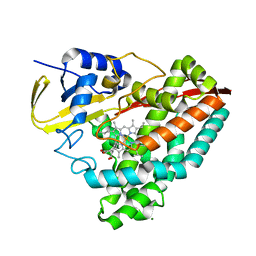 | |
5C86
 
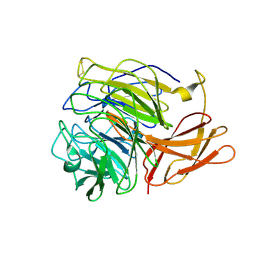 | | Novel fungal alcohol oxidase with catalytic diversity among the AA5 family, apo form | | Descriptor: | Kelch domain-containing protein | | Authors: | Urresti, S, Yin, D.T, LaFond, M, Derikvand, F, Berrin, G.J, Henrissat, B, Walton, P.H, Brumer, H, Davies, G.J. | | Deposit date: | 2015-06-25 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family.
Nat Commun, 6, 2015
|
|
7M5Z
 
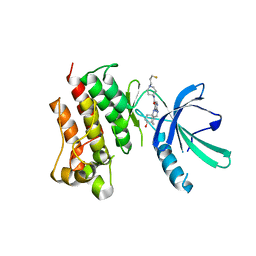 | | Crystal Structure of the MerTK Kinase Domain in Complex with Inhibitor MIPS15692 | | Descriptor: | 2-(butylamino)-N-[1-(3-fluoropropyl)piperidin-4-yl]-4-{[(1r,4r)-4-hydroxycyclohexyl]amino}pyrimidine-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Hermans, S.J, Hancock, N.C, Baell, J.B, Parker, M.W. | | Deposit date: | 2021-03-25 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Development of [ 18 F]MIPS15692, a radiotracer with in vitro proof-of-concept for the imaging of MER tyrosine kinase (MERTK) in neuroinflammatory disease.
Eur.J.Med.Chem., 226, 2021
|
|
8TD8
 
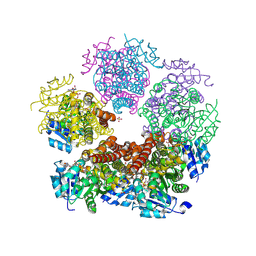 | | Structure of PYCR1 complexed with NADH and 2S-Hydroxy-3,3-dimethylbutyric acid | | Descriptor: | (2S)-2-hydroxy-3,3-dimethylbutanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
6MA2
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor ent-1a | | Descriptor: | Host Cell Factor 1 peptide, N-[(2S)-2-(2-methoxyphenyl)-2-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
