3MH2
 
 | | Mutagenesis of p38 MAP kinase establishes key roles of Phe169 in function and structural dynamics and reveals a novel DFG-out state | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Namboodiri, H.V, Karpusas, M, Bukhtiyarova, M, Springman, E.B. | | Deposit date: | 2010-04-07 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutagenesis of p38alpha MAP kinase establishes key roles of Phe169 in function and structural dynamics and reveals a novel DFG-OUT state.
Biochemistry, 46, 2007
|
|
7CXD
 
 | | Xray structure of rat Galectin-3 CRD in complex with TD-139 belonging to P121 space group | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, BROMIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
6S0Z
 
 | | Erythromycin Resistant Staphylococcus aureus 50S ribosome (delta R88 A89 uL22) in complex with erythromycin. | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Halfon, Y, Matozv, D, Eyal, Z, Bashan, A, Zimmerman, E, Kjeldgaard, J, Ingmer, H, Yonath, A. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant.
Sci Rep, 9, 2019
|
|
5IOU
 
 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the glutamate/glycine-bound conformation | | Descriptor: | GLUTAMIC ACID, GLYCINE, Ionotropic glutamate receptor subunit NR2B, ... | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IPU
 
 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 6 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
7CYY
 
 | | Crystal structure of Arabinose isomerase from hybrid AI8 with Adonitol | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, D-ribitol, L-arabinose isomerase, ... | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-09-05 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic hybrid AI8 with Adonitol
To Be Published
|
|
7CZJ
 
 | |
3MMC
 
 | | Structure of the dissimilatory sulfite reductase from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Schiffer, A, Parey, K, Warkentin, E, Diederichs, K, Huber, H, Stetter, K.O, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the dissimilatory sulfite reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Mol.Biol., 379, 2008
|
|
7CXO
 
 | | Crystal structure of Arabinose isomerase from hybrid AI10 | | Descriptor: | GLYCEROL, L-arabinose isomerase, MANGANESE (II) ION | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-09-02 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic hybrid AI10
To Be Published
|
|
7DRP
 
 | | Structure of ATP-grasp ligase PsnB complexed with phosphomimetic variant of minimal precursor, Mg, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-grasp domain-containing protein, MAGNESIUM ION, ... | | Authors: | Song, I, Yu, J, Song, W, Kim, S. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Molecular mechanism underlying substrate recognition of the peptide macrocyclase PsnB.
Nat.Chem.Biol., 17, 2021
|
|
7CZC
 
 | | Crystal structure of apo-FabG from Vibrio harveyi | | Descriptor: | 3-oxoacyl-ACP reductase FabG, DI(HYDROXYETHYL)ETHER | | Authors: | Singh, B.K, Kumar, A, Paul, B, Biswas, R, Das, A.K. | | Deposit date: | 2020-09-08 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of apo-FabG from Vibrio harveyi
To Be Published
|
|
7DRO
 
 | | Structure of ATP-grasp ligase PsnB complexed with minimal precursor | | Descriptor: | ATP-grasp domain-containing protein, PsnA214-38, Precursor peptide | | Authors: | Song, I, Yu, J, Song, W, Kim, S. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Molecular mechanism underlying substrate recognition of the peptide macrocyclase PsnB.
Nat.Chem.Biol., 17, 2021
|
|
7DRM
 
 | | Structure of ATP-grasp ligase PsnB complexed with minimal precursor, Mg, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-grasp domain-containing protein, MAGNESIUM ION, ... | | Authors: | Song, I, Yu, J, Song, W, Kim, S. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Molecular mechanism underlying substrate recognition of the peptide macrocyclase PsnB.
Nat.Chem.Biol., 17, 2021
|
|
3MRY
 
 | | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina with 6-aminopurine at 2.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of type I ribosome inactivating protein from Momordica balsamina with 6-aminopurine at 2.0A resolution
To be Published
|
|
7DRN
 
 | | Structure of ATP-grasp ligase PsnB complexed with precursor peptide PsnA2 and AMPPNP | | Descriptor: | ATP-grasp domain-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PsnA214-38, ... | | Authors: | Song, I, Yu, J, Song, W, Kim, S. | | Deposit date: | 2020-12-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Molecular mechanism underlying substrate recognition of the peptide macrocyclase PsnB.
Nat.Chem.Biol., 17, 2021
|
|
6RHZ
 
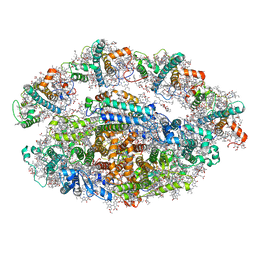 | | Structure of a minimal photosystem I from a green alga | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Perez Boerema, A, Klaiman, D, Caspy, I, Netzer-El, S.Y, Amunts, A, Nelson, N. | | Deposit date: | 2019-04-23 | | Release date: | 2020-02-19 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a minimal photosystem I from the green alga Dunaliella salina.
Nat.Plants, 6, 2020
|
|
7DDX
 
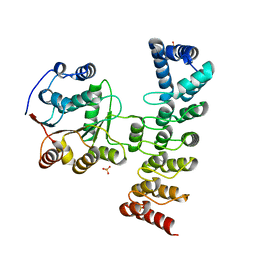 | | Crystal structure of KANK1 S1179F mutant in complex wtih eIF4A1 | | Descriptor: | Eukaryotic initiation factor 4A-I, GLYCEROL, KN motif and ankyrin repeat domains 1, ... | | Authors: | Pan, W, Xu, Y, Wei, Z. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nephrotic-syndrome-associated mutation of KANK2 induces pathologic binding competition with physiological interactor KIF21A.
J.Biol.Chem., 297, 2021
|
|
7DAJ
 
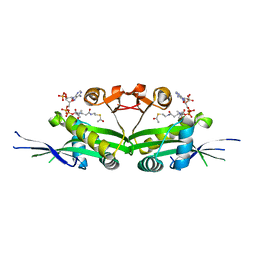 | | The crystal structure of serotonin N-acetyltransferase in complex with acetyl-CoA from Oryza Sativa | | Descriptor: | ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7D2F
 
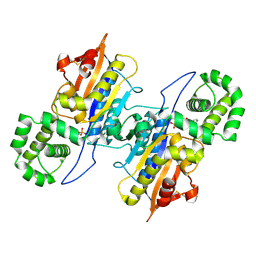 | | Lp major histidine acid phosphatase mutant D281A/5'-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Major acid phosphatase | | Authors: | Guo, Y, Teng, Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into a new substrate binding mode of a histidine acid phosphatase from Legionella pneumophila.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
7CYI
 
 | | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua | | Descriptor: | Alcohol dehydrogenase 1, ZINC ION | | Authors: | Feng, X, Fan, S, Lv, G, Li, M, Wu, G, Jin, Y, Yang, Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua
To Be Published
|
|
5I6X
 
 | | X-ray structure of the ts3 human serotonin transporter complexed with paroxetine at the central site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8B6 antibody, heavy chain, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
4PHF
 
 | | Crystal structure of Ypt7 covalently modified with GDP | | Descriptor: | GTP-binding protein YPT7, MAGNESIUM ION, N-[3-(propanoylamino)propyl]guanosine 5'-(trihydrogen diphosphate), ... | | Authors: | Vieweg, S, Wiegandt, D, Hofmann, F, Koch, D, Wu, Y, Itzen, A, Mueller, M.P, Goody, R.S. | | Deposit date: | 2014-05-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Locking GTPases covalently in their functional states.
Nat Commun, 6, 2015
|
|
7DHS
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1R)-1-phenylethyl]benzo[cd]indol-2-one, Bromodomain-containing protein 4 | | Authors: | Wu, T, Xiang, Q, Wang, C, Wu, C, Zhang, C, Zhang, M, Liu, Z, Zhang, Y, Xiao, L, Xu, Y. | | Deposit date: | 2020-11-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Y06014 is a selective BET inhibitor for the treatment of prostate cancer.
Acta Pharmacol.Sin., 42, 2021
|
|
7DJS
 
 | |
7DAI
 
 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa | | Descriptor: | Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
