1DT4
 
 | | CRYSTAL STRUCTURE OF NOVA-1 KH3 K-HOMOLOGY RNA-BINDING DOMAIN | | Descriptor: | NEURO-ONCOLOGICAL VENTRAL ANTIGEN 1 | | Authors: | Lewis, H.A, Chen, H, Edo, C, Buckanovich, R.J, Yang, Y.Y.L. | | Deposit date: | 2000-01-11 | | Release date: | 2000-02-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
4JT9
 
 | | Crystal Structure of human SIRT3 with ELT inhibitor 3 [14-(4-{2-[(methylsulfonyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide] | | Descriptor: | 4-(4-{2-[(methylsulfonyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide, GLYCEROL, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Dai, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery of Thieno[3,2-d]pyrimidine-6-carboxamides as Potent Inhibitors of SIRT1, SIRT2, and SIRT3.
J.Med.Chem., 56, 2013
|
|
3DN2
 
 | | Bromopentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1-bromo-2,3,4,5,6-pentafluorobenzene, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
3DN8
 
 | | Iodopentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant (seleno version) | | Descriptor: | 1,2,3,4,5-pentafluoro-6-iodobenzene, 2-HYDROXYETHYL DISULFIDE, BETA-MERCAPTOETHANOL, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
6KK7
 
 | | Structure of thermal-stabilised(M6) human GLP-1 receptor transmembrane domain | | Descriptor: | Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | Authors: | Song, G. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
5GK3
 
 | | Native structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.8 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fructose-bisphosphate aldolase class 2, GLYCEROL, ... | | Authors: | Tran, T.H, Huynh, K.H, Ho, T.H, Kang, L.W. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Apo structure of fructose 1,6-bisphosphate aldolase from Escherichia coli at 1.8 Angstrom resolution
To Be Published
|
|
7DCG
 
 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with maltotriose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3JWO
 
 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D Heavy CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Huang, C.C, Kwon, Y.D, Zhou, T, Robinson, J.E, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5C42
 
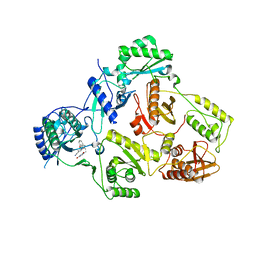 | | Crystal Structure of HIV-1 Reverse Transcriptase (K101P) Variant in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)indolizine-2-carbonitrile (JLJ555), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}indolizine-2-carbonitrile, HIV-1 Reverse Transcriptase, p51 subunit, ... | | Authors: | Frey, K.M, Gray, W.T, Anderson, K.S. | | Deposit date: | 2015-06-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent Inhibitors Active against HIV Reverse Transcriptase with K101P, a Mutation Conferring Rilpivirine Resistance.
Acs Med.Chem.Lett., 6, 2015
|
|
3DHA
 
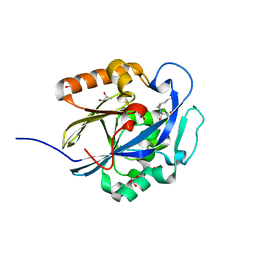 | | An Ultral High Resolution Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at An Alternative Site | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
4AHU
 
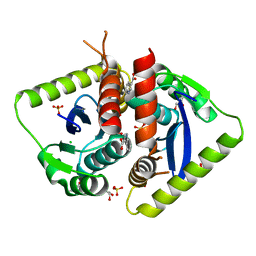 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOLE-3-CARBOXYLIC ACID, ACETIC ACID, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-07 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
7KR3
 
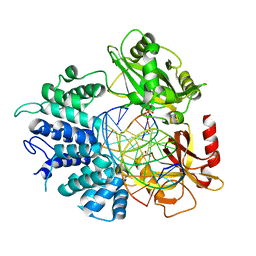 | |
4UIV
 
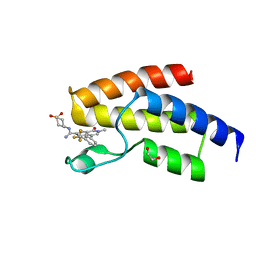 | | BROMODOMAIN OF HUMAN BRD9 WITH N-(1,1-dioxo-1-thian-4-yl)-5-methyl-4- oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2- carboximidamide | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN-CONTAINING PROTEIN 9, N-(1,1-dioxo-1-thian-4-yl)-5-methyl-4-oxo-7-3-(trifluoromethyl)phenyl-4H,5H-thieno-3,2-c-pyridine-2-carboximidamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
3FKH
 
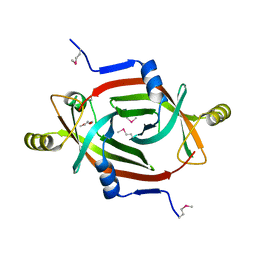 | |
4J5R
 
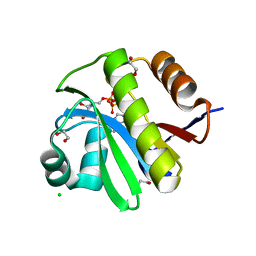 | | TARG1 (C6orf130), Terminal ADP-ribose Glycohydrolase 1 bound to ADP-HPD | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, CHLORIDE ION, ... | | Authors: | Schellenberg, M.J, Appel, C.D, Krahn, J, Williams, R.S. | | Deposit date: | 2013-02-09 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Embo J., 32, 2013
|
|
1L6Z
 
 | | CRYSTAL STRUCTURE OF MURINE CEACAM1A[1,4]: A CORONAVIRUS RECEPTOR AND CELL ADHESION MOLECULE IN THE CEA FAMILY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, biliary glycoprotein C | | Authors: | Tan, K, Zelus, B.D, Meijers, R, Liu, J.-H, Bergelson, J.M, Duke, N, Zhang, R, Joachimiak, A, Holmes, K.V, Wang, J.-H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | CRYSTAL STRUCTURE OF MURINE sCEACAM1a[1,4]: A CORONAVIRUS RECEPTOR IN THE CEA FAMILY
Embo J., 21, 2002
|
|
5UKZ
 
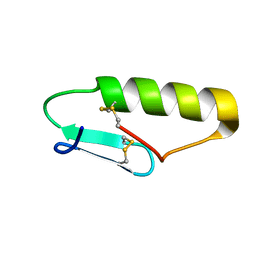 | | NMR Solution structure of chemically synthesized antilisterial Pediocin PA-1 M31L analog. | | Descriptor: | Bacteriocin pediocin PA-1 M31L | | Authors: | Bedard, F, Hammami, R, Zirah, S, Rebuffat, S, Fliss, I, Biron, E. | | Deposit date: | 2017-01-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Synthesis, antimicrobial activity and conformational analysis of the class IIa bacteriocin pediocin PA-1 and analogs thereof.
Sci Rep, 8, 2018
|
|
5URS
 
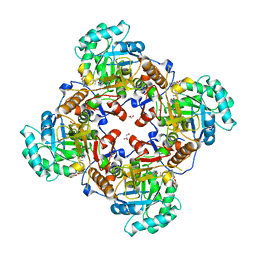 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P178 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-12 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P178
To Be Published
|
|
1SUX
 
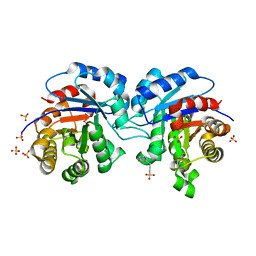 | | CRYSTALLOGRAPHIC ANALYSIS OF THE COMPLEX BETWEEN TRIOSEPHOSPHATE ISOMERASE FROM TRYPANOSOMA CRUZI AND 3-(2-benzothiazolylthio)-1-propanesulfonic acid | | Descriptor: | 3-(2-BENZOTHIAZOLYLTHIO)-1-PROPANESULFONIC ACID, SULFATE ION, Triosephosphate isomerase, ... | | Authors: | Tellez-Valencia, A, Olivares-Illana, V, Hernandez-Santoyo, A, Perez-Montfort, R, Costas, M, Rodriguez-Romero, A, Tuena De Gomez-Puyou, M, Gomez-Puyou, A. | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inactivation of triosephosphate isomerase from Trypanosoma cruzi by an agent that perturbs its dimer interface.
J.Mol.Biol., 341, 2004
|
|
2WMT
 
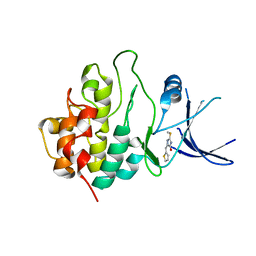 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 2-(methylsulfanyl)-5-(thiophen-2-ylmethyl)-1H-imidazol-4-ol, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
7FZ5
 
 | | Crystal Structure of human FABP4 in complex with rac-(2R)-1-[[3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl]pyrrolidine-2-carboxylic acid | | Descriptor: | 1-{[(3M)-3-(3-cyclopropyl-1,2,4-oxadiazol-5-yl)-4,5,6,7-tetrahydro-1-benzothiophen-2-yl]carbamoyl}-D-proline, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Neidhart, W, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
4HHL
 
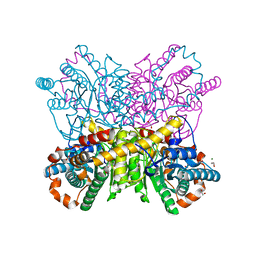 | | High resolution crystal structure of Glucose Isomerase from Streptomyces sp. SK | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Ben Hlima, H, Riguet, J, Haser, R, Aghajari, N. | | Deposit date: | 2012-10-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of critical residues for the activity and thermostability of Streptomyces sp. SK glucose isomerase.
Appl.Microbiol.Biotechnol., 97, 2013
|
|
5G1D
 
 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | Descriptor: | SYNDECAN-4, SYNTENIN-1 | | Authors: | Lee, I, Kim, H, Yun, J.H, Lee, W. | | Deposit date: | 2016-03-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
5G1E
 
 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | Descriptor: | SYNTENIN-1 | | Authors: | Lee, I, Kim, H, Yun, J.H, Lee, W. | | Deposit date: | 2016-03-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
4AAC
 
 | | P38ALPHA MAP KINASE BOUND TO CMPD 29 | | Descriptor: | CHLORIDE ION, MITOGEN-ACTIVATED PROTEIN KINASE 14, N-isoxazol-3-yl-4-methyl-3-[6-(4-methylpiperazin-1-yl)-4-oxo-quinazolin-3-yl]benzamide | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
