5Z4A
 
 | | Structure of Tailor in complex with AGU RNA | | Descriptor: | RNA (5'-R(*AP*GP*U)-3'), Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-10 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.637 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
6A4W
 
 | | AcrR from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Kang, S.M, Kim, D.H. | | Deposit date: | 2018-06-21 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | The crystal structure of AcrR from Mycobacterium tuberculosis reveals a one-component transcriptional regulation mechanism.
Febs Open Bio, 9, 2019
|
|
6AJM
 
 | | Crystal structure of apo AtaTR | | Descriptor: | DUF1778 domain-containing protein, N-acetyltransferase, TRIETHYLENE GLYCOL | | Authors: | Yashiro, Y, Yamashita, S, Tomita, K. | | Deposit date: | 2018-08-28 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal Structure of the Enterohemorrhagic Escherichia coli AtaT-AtaR Toxin-Antitoxin Complex.
Structure, 27, 2019
|
|
6E1A
 
 | | Menin bound to M-89 | | Descriptor: | (1S,2R)-2-[(4S)-2-methyl-4-{1-[(1-{4-[(pyridin-4-yl)sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}-1,2,3,4-tetrahydroisoquinolin-4-yl]cyclopentyl methylcarbamate, Menin, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2018-07-09 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery of M-89 as a Highly Potent Inhibitor of the Menin-Mixed Lineage Leukemia (Menin-MLL) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
2IF5
 
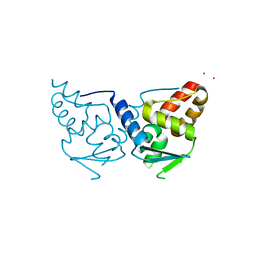 | | Structure of the POZ domain of human LRF, a master regulator of oncogenesis | | Descriptor: | PRASEODYMIUM ION, Zinc finger and BTB domain-containing protein 7A | | Authors: | Schubot, F.D, Waugh, D.S, Tropea, J. | | Deposit date: | 2006-09-20 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the POZ domain of human LRF, a master regulator of oncogenesis.
Biochem.Biophys.Res.Commun., 351, 2006
|
|
6RX9
 
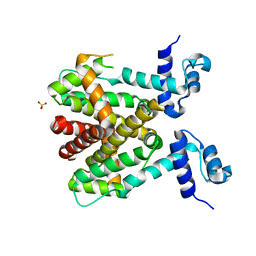 | |
6RXB
 
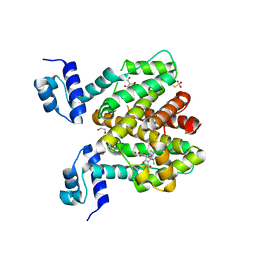 | | Crystal structure of TetR-Q116A from Acinetobacter baumannii AYE in complex with minocycline | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tam, H.K, Himpich, S, Pos, K.M. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding of Tetracyclines to Acinetobacter baumannii TetR Involves Two Arginines as Specificity Determinants
Front Microbiol, 2021
|
|
5U8M
 
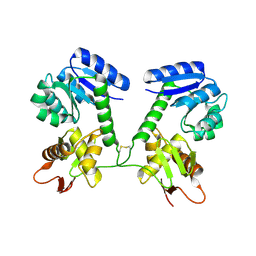 | |
3PM1
 
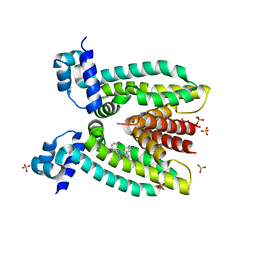 | | Structure of QacR E90Q bound to Ethidium | | Descriptor: | ETHIDIUM, HTH-type transcriptional regulator qacR, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A single acidic residue can guide binding site selection but does not govern QacR cationic-drug affinity.
Plos One, 6, 2011
|
|
4CRO
 
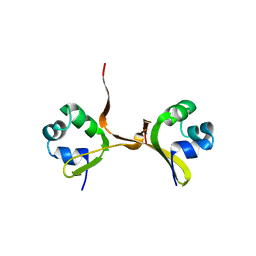 | | PROTEIN-DNA CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURE OF A LAMBDA CRO-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*TP*CP*AP*CP*CP*GP*CP*GP*GP*GP*TP*GP*AP*TP*A)-3'), PROTEIN (LAMBDA CRO) | | Authors: | Brennan, R.G, Roderick, S.L, Takeda, Y, Matthews, B.W. | | Deposit date: | 1992-01-15 | | Release date: | 1992-01-15 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Protein-DNA conformational changes in the crystal structure of a lambda Cro-operator complex.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
2GPE
 
 | |
5VFA
 
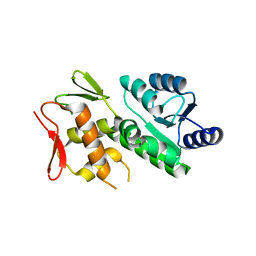 | | RitR Mutant - C128D | | Descriptor: | Response regulator | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2017-04-07 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | RitR is an archetype for a novel family of redox sensors in the streptococci that has evolved from two-component response regulators and is required for pneumococcal colonization.
PLoS Pathog., 14, 2018
|
|
6C3N
 
 | | Crystal structure of BCL6 BTB domain in complex with compound 7CC5 | | Descriptor: | B-cell lymphoma 6 protein, N-(2-phenylethyl)-N'-pyridin-3-ylthiourea | | Authors: | Linhares, B, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53170586 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
5NAF
 
 | | Co-crystal structure of an MeCP2 peptide with TBLR1 WD40 domain | | Descriptor: | F-box-like/WD repeat-containing protein TBL1XR1, GLYCEROL, Methyl-CpG-binding protein 2 | | Authors: | Kruusvee, V, Cook, A.G. | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structure of the MeCP2-TBLR1 complex reveals a molecular basis for Rett syndrome and related disorders.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7B0C
 
 | | [4Fe-4S]-NsrR complexed to 23-bp HmpA1 operator fragment | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*GP*AP*AP*TP*AP*TP*CP*AP*TP*CP*TP*AP*CP*CP*AP*AP*TP*T)-3'), DNA (5'-D(P*AP*AP*TP*TP*GP*GP*TP*AP*GP*AP*TP*GP*AP*TP*AP*TP*TP*CP*GP*TP*GP*TP*T)-3'), HTH-type transcriptional repressor NsrR, ... | | Authors: | Rohac, R, Fontecilla-Camps, J.C, Volbeda, A. | | Deposit date: | 2020-11-19 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural determinants of DNA recognition by the NO sensor NsrR and related Rrf2-type [FeS]-transcription factors.
Commun Biol, 5, 2022
|
|
6C3L
 
 | | Crystal structure of BCL6 BTB domain with compound 15f | | Descriptor: | B-cell lymphoma 6 protein, N-[2-(1H-indol-3-yl)ethyl]-N'-{3-[(4-methylpiperazin-1-yl)methyl]-1-[2-(morpholin-4-yl)-2-oxoethyl]-1H-indol-6-yl}thiourea | | Authors: | Linhares, B, Cheng, H, Cierpicki, T, Xue, F. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.46092153 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
4UV8
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Benzyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [[(2R,3S,4S)-5-[(4aS,10aS)-4a-[(1S)-3-azanylidene-1,4-diphenyl-butyl]-7,8-dimethyl-2,4-bis(oxidanylidene)-5,10a-dihydro-1H-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentoxy]-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
6CQ1
 
 | | BCL6 BTB domain in complex with 15a | | Descriptor: | 2-{6-({[2-(1H-indol-3-yl)ethyl]carbamothioyl}amino)-3-[(4-methylpiperazin-1-yl)methyl]-1H-indol-1-yl}-N-(propan-2-yl)acetamide, B-cell lymphoma 6 protein | | Authors: | Linhares, B.M, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69921041 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
2HSG
 
 | |
7M10
 
 | | PHF2 PHD Domain Complexed with Peptide From N-terminus of VRK1 | | Descriptor: | FORMIC ACID, Lysine-specific demethylase PHF2, Serine/threonine-protein kinase VRK1 N-terminus peptide, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2021-03-11 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Histone H3 N-terminal mimicry drives a novel network of methyl-effector interactions.
Biochem.J., 478, 2021
|
|
2ISY
 
 | | Crystal structure of the nickel-activated two-domain iron-dependent regulator (IdeR) | | Descriptor: | Iron-dependent repressor ideR, NICKEL (II) ION, PHOSPHATE ION | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
6KR6
 
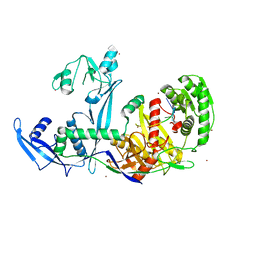 | | Crystal structure of Drosophila Piwi | | Descriptor: | MERCURY (II) ION, Protein piwi, ZINC ION, ... | | Authors: | Yamaguchi, S, Oe, A, Yamashita, K, Hirano, S, Mastumoto, N, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Drosophila Piwi.
Nat Commun, 11, 2020
|
|
2ISZ
 
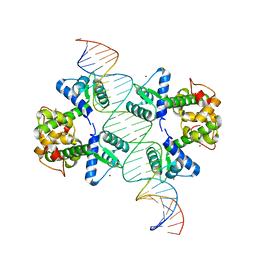 | | Crystal structure of a two-domain IdeR-DNA complex crystal form I | | Descriptor: | Iron-dependent repressor ideR, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
2IT0
 
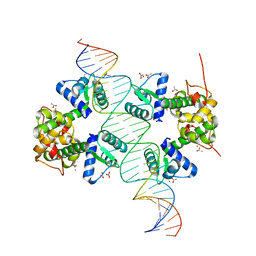 | | Crystal structure of a two-domain IdeR-DNA complex crystal form II | | Descriptor: | ACETATE ION, Iron-dependent repressor ideR, NICKEL (II) ION, ... | | Authors: | Wisedchaisri, G, Chou, C.J, Wu, M, Roach, C, Rice, A.E, Holmes, R.K, Beeson, C, Hol, W.G. | | Deposit date: | 2006-10-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures, metal activation, and DNA-binding properties of two-domain IdeR from Mycobacterium tuberculosis
Biochemistry, 46, 2007
|
|
4XTU
 
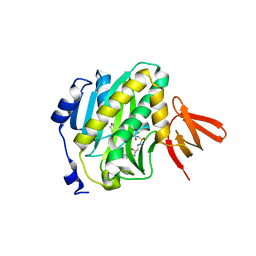 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor (N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide) | | Descriptor: | Bifunctional ligase/repressor BirA, N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6500299 Å) | | Cite: | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor (N-({[(1R,2S,3R,4R)-4-(6-amino-9H-purin-9-yl)-2,3-dihydroxycyclopentyl]methyl}sulfamoyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide)
To be Published
|
|
