4P0N
 
 | | Crystal structure of PDE10a with a novel Imidazo[4,5-b]pyridine inhibitor | | Descriptor: | GLYCEROL, N-[cis-3-(2-methoxy-3H-imidazo[4,5-b]pyridin-3-yl)cyclobutyl]-1,3-benzothiazol-2-amine, N-[trans-3-(2-methoxy-3H-imidazo[4,5-b]pyridin-3-yl)cyclobutyl]-1,3-benzothiazol-2-amine, ... | | Authors: | Chmait, S. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Novel Imidazo[4,5-b]pyridines as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
Acs Med.Chem.Lett., 5, 2014
|
|
3A1E
 
 | | Crystal structure of the P- and N-domains of His462Gln mutant CopA, a copper-transporting P-type ATPase, bound with AMPPCP-Mg | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Probable copper-exporting P-type ATPase A | | Authors: | Tsuda, T, Toyoshima, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Nucleotide recognition by CopA, a Cu+-transporting P-type ATPase.
Embo J., 28, 2009
|
|
4MUW
 
 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | Descriptor: | 2-{4-[(6,7-difluoro-1H-benzimidazol-2-yl)amino]phenoxy}-N-methyl-3,4'-bipyridin-2'-amine, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S, Jordan, S. | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
7PJI
 
 | |
8WDO
 
 | | Crystal structure of PDE4D complexed with DCN | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-2-(3,4-dimethoxyphenyl)-3-(hydroxymethyl)-7-methoxy-2,3-dihydro-1-benzofuran-5-yl]propan-1-ol, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of Dihydrobenzofuran Neolignans as Novel PDE4 Inhibitors and Evaluation of Antiatopic Dermatitis Efficacy in DNCB-Induced Mice Model.
J.Med.Chem., 67, 2024
|
|
8WDN
 
 | | Crystal structure of PDE4D complexed with 7b-1 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(2~{S},3~{R})-7-ethoxy-2-(3-ethoxy-4-methoxy-phenyl)-3-(hydroxymethyl)-2,3-dihydro-1-benzofuran-5-yl]propan-1-ol, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-09-15 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of Dihydrobenzofuran Neolignans as Novel PDE4 Inhibitors and Evaluation of Antiatopic Dermatitis Efficacy in DNCB-Induced Mice Model.
J.Med.Chem., 67, 2024
|
|
6SWY
 
 | | Structure of active GID E3 ubiquitin ligase complex minus Gid2 and delta Gid9 RING domain | | Descriptor: | Glucose-induced degradation protein 8, Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-09-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly
Mol.Cell, 77, 2020
|
|
2Y0J
 
 | | Triazoloquinazolines as a novel class of phosphodiesterase 10A (PDE10A) inhibitors, part 2, Lead-optimisation. | | Descriptor: | 5-(1H-BENZIMIDAZOL-2-YLMETHYLSULFANYL)-2-METHYL-[1,2,4]TRIAZOLO[1,5-C]QUINAZOLINE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Kehler, J, Ritzen, A, Langgard, M, Petersen, S.L, Christoffersen, C.T, Nielsen, J, Kilburn, J.P. | | Deposit date: | 2010-12-03 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Triazoloquinazolines as a Novel Class of Phosphodiesterase 10A (Pde10A) Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8G38
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
6JIC
 
 | | Identification and Characterization of a carboxypeptidase inhibitor from Lycium barbarum | | Descriptor: | WCI | | Authors: | Tan, W.L, Wong, K.H, Huang, J.Y, Tay, S.V, Wang, S.J. | | Deposit date: | 2019-02-20 | | Release date: | 2020-02-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and characterization of a wolfberry carboxypeptidase inhibitor from Lycium barbarum.
Food Chem, 351, 2021
|
|
7RD1
 
 | | The Capsid Structure of the ChAdOx1 viral vector/chimpanzee adenovirus Y25 | | Descriptor: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | Authors: | Baker, A.T, Boyd, R.J, Sarkar, D, Vermaas, J.V, Williams, D, Singharoy, A. | | Deposit date: | 2021-07-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | ChAdOx1 interacts with CAR and PF4 with implications for thrombosis with thrombocytopenia syndrome.
Sci Adv, 7, 2021
|
|
2I3V
 
 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of G725C mutant | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
3UG2
 
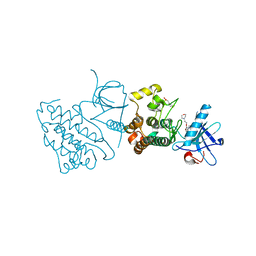 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with gefitinib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, Gefitinib | | Authors: | Parker, L.J, Handa, N, Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 32, 2013
|
|
6T8H
 
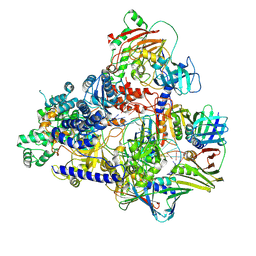 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | Descriptor: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
1F9H
 
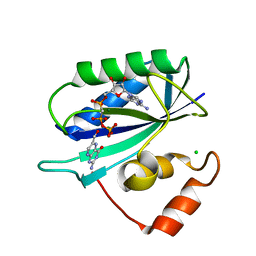 | | CRYSTAL STRUCTURE OF THE TERNARY COMPLEX OF E. COLI HPPK(R92A) WITH MGAMPCPP AND 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 1.50 ANGSTROM RESOLUTION | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2000-07-10 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dynamic Roles of Arginine Residues 82 and 92 of Escherichia coli
6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase: Crystallographic
Studies
Biochemistry, 42, 2003
|
|
2E1Y
 
 | |
2E20
 
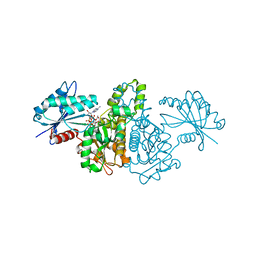 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with diadenosine tetraphosphate (Ap4A) | | Descriptor: | 1,2-ETHANEDIOL, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Propionate Kinase | | Authors: | Simanshu, D.K, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2006-11-04 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Salmonella typhimurium propionate kinase and its complex with Ap4A: evidence for a novel Ap4A synthetic activity.
Proteins, 70, 2008
|
|
3UG1
 
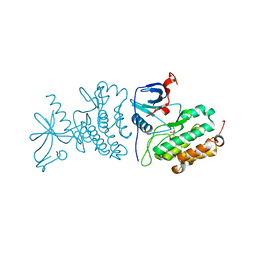 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in the apo form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor | | Authors: | Parker, L.J, Handa, N, Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 32, 2013
|
|
4OGB
 
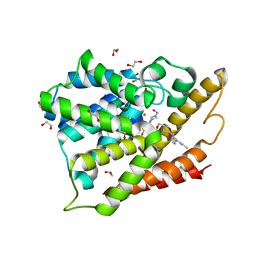 | | Crystal structure of the catalytic domain of PDE4D2 with compound 2 | | Descriptor: | (2R)-8-(3,4-dimethoxyphenyl)-6-methyl-2-(tetrahydro-2H-pyran-4-yl)-2H-chromen-4-ol, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Feil, S.C, Parker, M.W. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | The PDE inhibition profile of LY294002 and tetrahydropyranyl analogues reveals a chromone motif for the development of PDE inhibitors
To be Published
|
|
3W5E
 
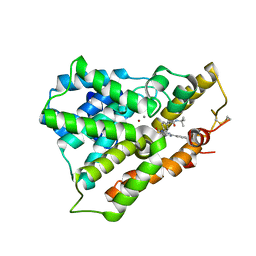 | | Crystal structure of phosphodiesterase 4B in complex with compound 31e | | Descriptor: | CALCIUM ION, N-tert-butyl-2-{4-[(5,5-dioxido-2-phenyl-7,8-dihydro-6H-thiopyrano[3,2-d]pyrimidin-4-yl)amino]phenyl}acetamide, ZINC ION, ... | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2013-01-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of the fused bicyclic 4-amino-2-phenylpyrimidine derivatives as novel and potent PDE4 inhibitors
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6GNZ
 
 | |
4P1R
 
 | |
2E3C
 
 | |
4PHW
 
 | |
3WD9
 
 | | Crystal structure of phosphodiesterase 4B in complex with compound 10f | | Descriptor: | 4-[(4-{2-[(2,2-dimethylpropyl)amino]-2-oxoethyl}phenyl)amino]-2-phenylpyrimidine-5-carboxamide, CALCIUM ION, ZINC ION, ... | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2013-06-11 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and biological evaluation of 5-carbamoyl-2-phenylpyrimidine derivatives as novel and potent PDE4 inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
