5DZ5
 
 | | Crystal structure of Dendroaspis polylepis mambalgin-1 wild-type in P41212 space group | | Descriptor: | Mambalgin-1 | | Authors: | Stura, E.A, Tepshi, L, Mourier, G, Kessler, P, Servent, D. | | Deposit date: | 2015-09-25 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition.
J.Biol.Chem., 291, 2016
|
|
5JYN
 
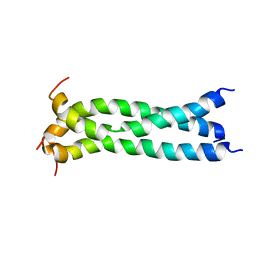 | | Structure of the transmembrane domain of HIV-1 gp41 in bicelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Dev, J, Fu, Q, Park, D, Chen, B, Chou, J.J. | | Deposit date: | 2016-05-14 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for membrane anchoring of HIV-1 envelope spike.
Science, 353, 2016
|
|
7TCA
 
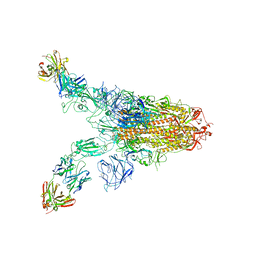 | | Cryo-EM structure of SARS-CoV-2 Omicron spike in complex with antibody A19-46.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody A19-46.1, ... | | Authors: | Zhou, T, Kwong, P.D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
4AZI
 
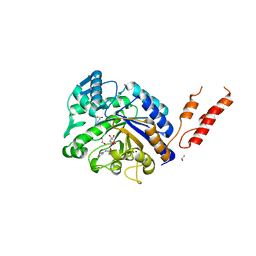 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE, BICINE, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
6BUL
 
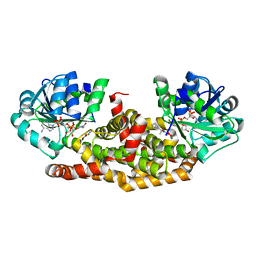 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 2 | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McFeary, R.P. | | Deposit date: | 2017-12-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductoisomerase (KARI) inhibitors
To Be Published
|
|
7YJS
 
 | |
1SZS
 
 | | The structure of gamma-aminobutyrate aminotransferase mutant: I50Q | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
5AGR
 
 | | Crystal structure of the LeuRS editing domain of Mycobacterium tuberculosis in complex with the adduct (S)-3-(Aminomethyl)-7-ethoxybenzo[c][1,2]oxaborol-1(3H)-ol-AMP | | Descriptor: | 1,2-ETHANEDIOL, 3-AMINOMETHYL-7-(ETHOXY)-3H-BENZO[C][1,2]OXABOROL-1-OL modified adenosine, LEUCINE, ... | | Authors: | Palencia, A, Li, X, Alley, M.R.K, Ding, C, Easom, E.E, Hernandez, V, Meewan, M, Mohan, M, Rock, F.L, Franzblau, S.G, Wang, Y, Lenaerts, A.J, Parish, T, Cooper, C.B, Waters, M.G, Ma, Z, Mendoza, A, Barros, D, Cusack, S, Plattner, J.J. | | Deposit date: | 2015-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Novel Oral Protein Synthesis Inhibitors of Mycobacterium Tuberculosis that Target Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
5NN5
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with 1-deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
9EBO
 
 | | Peptide 2 (GLP-1 (ACPC18)) bound to GLP-1R/Gs complex (conformer 1) | | Descriptor: | Glucagon-like peptide 1 receptor, Glucagon-like peptide 1(7-36), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cary, B.P, Hager, M.V, Mariam, Z, Morris, R.K, Belousoff, M.J, Deganutti, G, Wootten, D, Sexton, P.M, Gellman, S.H. | | Deposit date: | 2024-11-12 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Prolonged signaling of backbone-modified glucagon-like peptide- 1 analogues with diverse receptor trafficking.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8BCV
 
 | | Photosystem I assembly intermediate of Avena sativa | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A, Nelson, N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Photosystem I assembly intermediate of Avena sativa
To Be Published
|
|
2VCE
 
 | | Characterization and engineering of the bifunctional N- and O- glucosyltransferase involved in xenobiotic metabolism in plants | | Descriptor: | 1,2-ETHANEDIOL, 2,4,5-trichlorophenol, HYDROQUINONE GLUCOSYLTRANSFERASE, ... | | Authors: | Brazier-Hicks, M, Offen, W.A, Gershater, M.C, Revett, T.J, Lim, E.K, Bowles, D.J, Davies, G.J, Edwards, R. | | Deposit date: | 2007-09-20 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and Engineering of the Bifunctional N- and O-Glucosyltransferase Involved in Xenobiotic Metabolism in Plants.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
6DCW
 
 | |
6DD5
 
 | | Crystal Structure of the Cas6 Domain of Marinomonas mediterranea MMB-1 Cas6-RT-Cas1 Fusion Protein | | Descriptor: | GLYCEROL, MMB-1 Cas6 Fused to Maltose Binding Protein,CRISPR-associated endonuclease Cas1, SULFATE ION, ... | | Authors: | Stamos, J.L, Mohr, G, Silas, S, Makarova, K.S, Markham, L.M, Yao, J, Lucas-Elio, P, Sanchez-Amat, A, Fire, A.Z, Koonin, E.V, Lambowitz, A.M. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition.
Mol. Cell, 72, 2018
|
|
7BGI
 
 | | Photosystem I of a temperature sensitive mutant Chlamydomonas reinhardtii | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Caspy, I, Nelson, N. | | Deposit date: | 2021-01-07 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant
Commun Biol, 4, 2021
|
|
7BLX
 
 | | Photosystem I of a temperature sensitive mutant Chlamydomonas reinhardtii | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Caspy, I, Nelson, N. | | Deposit date: | 2021-01-19 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Dimeric and high-resolution structures of Chlamydomonas Photosystem I from a temperature-sensitive Photosystem II mutant
Commun Biol, 4, 2021
|
|
6RPG
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with inhibitor | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[2-[[4-[[3-[3-[[4-[(2-acetamidoethylamino)methyl]-5-[(5-cyanopyridin-3-yl)methoxy]-2-methyl-phenoxy]methyl]-2-methyl-phenyl]-2-methyl-phenyl]methoxy]-2-[(5-cyanopyridin-3-yl)methoxy]-5-methyl-phenyl]methylamino]ethyl]ethanamide | | Authors: | Magiera-Mularz, K, Basu, S, Yang, J, Xu, B, Skalniak, L, Musielak, B, Kholodovych, V, Holak, T.A, Hu, L. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, Evaluation, and Structural Studies ofC2-Symmetric Small Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6U23
 
 | | EM structure of MPEG-1(w.t.) soluble pre-pore | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-19 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
6MA8
 
 | | Human CYP3A4 bound to PMSF | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450 3A4, DIMETHYL SULFOXIDE, ... | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-08-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Interaction of Human Drug-Metabolizing CYP3A4 with Small Inhibitory Molecules.
Biochemistry, 58, 2019
|
|
5L6S
 
 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a positive allosteric regulator beta-fructose-1,6-diphosphate (FBP) - AGPase*FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
7DV0
 
 | |
7DV1
 
 | |
7DUZ
 
 | |
5ZH9
 
 | |
4YWT
 
 | |
