6SIQ
 
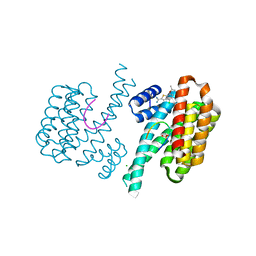 | | Fragment AZ-012 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-chloranyl-7-propan-2-yloxy-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Leysen, S, Wolter, M, Guillory, X, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-08-10 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.60121 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
3CGI
 
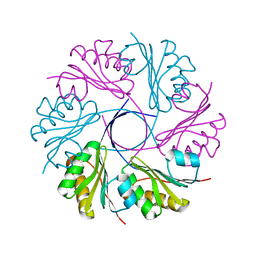 | |
5FAN
 
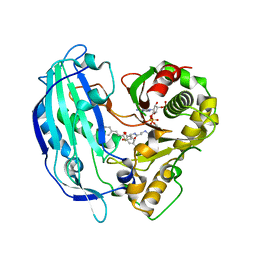 | | Crystal structure of PvHCT in complex with p-coumaroyl-CoA and protocatechuate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyltransferase 2, p-coumaroyl-CoA | | Authors: | Pereira, J.H, Moriarty, N.W, Eudes, A, Yogiswara, S, Wang, G, Benites, V.T, Baidoo, E.E.K, Lee, T.S, Keasling, J.D, Loque, D, Adams, P.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Exploiting the Substrate Promiscuity of Hydroxycinnamoyl-CoA:Shikimate Hydroxycinnamoyl Transferase to Reduce Lignin.
Plant Cell.Physiol., 57, 2016
|
|
3EZI
 
 | |
4JBQ
 
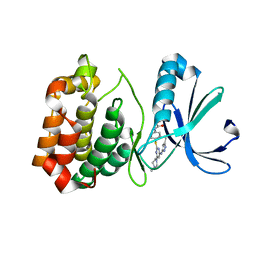 | | Novel Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules | | Descriptor: | Aurora Kinase A, CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE | | Authors: | Wu, J.S, Leou, J.S, Peng, Y.H, Hsueh, C.C, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2013-02-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aurora kinase inhibitors reveal mechanisms of HURP in nucleation of centrosomal and kinetochore microtubules.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6S4D
 
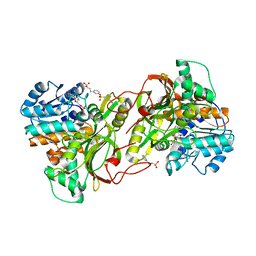 | |
1Y20
 
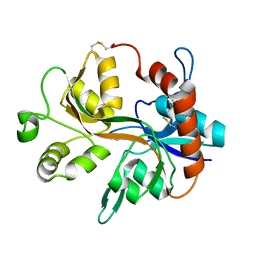 | |
5UR7
 
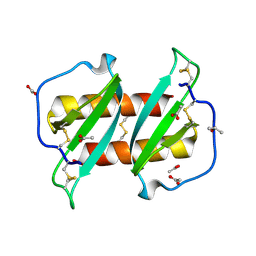 | |
1ZP0
 
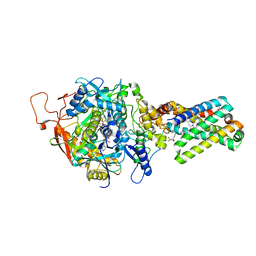 | | Crystal Structure of Mitochondrial Respiratory Complex II bound with 3-nitropropionate and 2-thenoyltrifluoroacetone | | Descriptor: | 3-NITROPROPANOIC ACID, 4,4,4-TRIFLUORO-1-THIEN-2-YLBUTANE-1,3-DIONE, FAD-binding protein, ... | | Authors: | Sun, F, Huo, X, Zhai, Y, Wang, A, Xu, J, Su, D, Bartlam, M, Rao, Z. | | Deposit date: | 2005-05-16 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Mitochondrial Respiratory Membrane Protein Complex II
Cell(Cambridge,Mass.), 121, 2005
|
|
5IGJ
 
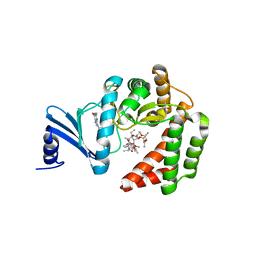 | |
2A6S
 
 | |
3SZB
 
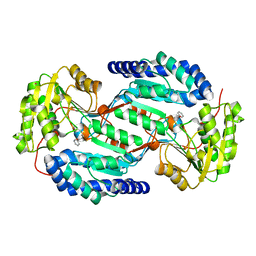 | |
5D6U
 
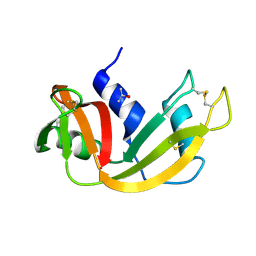 | |
1JW6
 
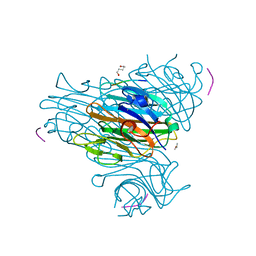 | | Crystal Structure of the Complex of Concanavalin A and Hexapeptide | | Descriptor: | CALCIUM ION, Concanavalin A, ISOPROPYL ALCOHOL, ... | | Authors: | Zhang, Z, Qian, M, Huang, Q, Jia, Y, Tang, Y. | | Deposit date: | 2001-09-02 | | Release date: | 2001-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the complex of concanavalin A and hexapeptide.
J.Protein Chem., 20, 2001
|
|
5D97
 
 | | Neutron crystal structure of H2O-solvent ribonuclease A | | Descriptor: | ISOPROPYL ALCOHOL, Ribonuclease pancreatic | | Authors: | Chatake, T, Fujiwara, S. | | Deposit date: | 2015-08-18 | | Release date: | 2016-04-06 | | Last modified: | 2018-12-05 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | A technique for determining the deuterium/hydrogen contrast map in neutron macromolecular crystallography
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5IGI
 
 | |
5IGP
 
 | |
8JY7
 
 | |
6FUE
 
 | | Periplasmic coiled coil domain of the FapF amyloid transporter | | Descriptor: | FapF, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Rouse, S.L, Stylianou, F, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-02-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The FapF Amyloid Secretion Transporter Possesses an Atypical Asymmetric Coiled Coil.
J. Mol. Biol., 430, 2018
|
|
1AIM
 
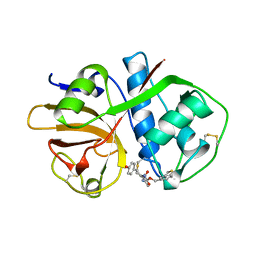 | |
5TVO
 
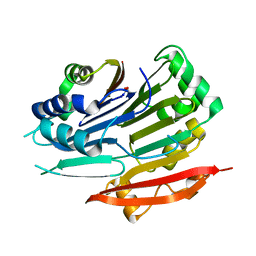 | | Crystal structure of Trypanosoma brucei AdoMetDC-delta26 monomer | | Descriptor: | PYRUVIC ACID, S-adenosylmethionine decarboxylase proenzyme, SODIUM ION | | Authors: | Volkov, O.A, Ariagno, C, Chen, Z, Tomchick, D.R, Phillips, M.A. | | Deposit date: | 2016-11-09 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Relief of autoinhibition by conformational switch explains enzyme activation by a catalytically dead paralog.
Elife, 5, 2016
|
|
6G1B
 
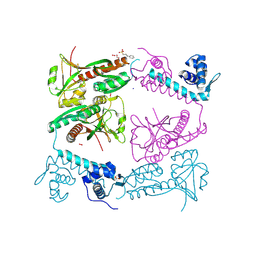 | | Corynebacterium glutamicum OxyR, oxidized form | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, FORMIC ACID, HYDROGEN PEROXIDE, ... | | Authors: | Young, D.R, Pedre, B.P, Messens, J.M. | | Deposit date: | 2018-03-21 | | Release date: | 2018-12-05 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1SZR
 
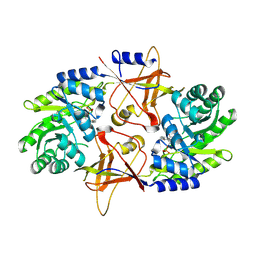 | | A Dimer interface mutant of ornithine decarboxylase reveals structure of gem diamine intermediate | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N~2~-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-ORNITHINE, Ornithine decarboxylase, ... | | Authors: | Jackson, L.K, Baldwin, J, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2004-04-06 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Multiple active site conformations revealed by distant site mutation in ornithine decarboxylase
Biochemistry, 43, 2004
|
|
5PHM
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D in complex with N09455a | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, N-OXALYLGLYCINE, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PHD
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D in complex with N09419a | | Descriptor: | 1,2-ETHANEDIOL, 4-NITROCATECHOL, Lysine-specific demethylase 4D, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.359 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
