3JRD
 
 | |
4GCT
 
 | | structure of No factor protein-DNA complex | | Descriptor: | DNA (5'-D(*TP*TP*AP*CP*GP*TP*GP*AP*GP*TP*AP*CP*TP*CP*AP*CP*GP*TP*AP*A)-3'), Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-07-30 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GCK
 
 | | structure of no-dna complex | | Descriptor: | DNA (5'-D(*GP*TP*GP*AP*GP*TP*AP*CP*TP*CP*AP*C)-3'), Nucleoid occlusion factor SlmA | | Authors: | Schumacher, M.A. | | Deposit date: | 2012-07-30 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1WID
 
 | | Solution Structure of the B3 DNA-Binding Domain of RAV1 | | Descriptor: | DNA-binding protein RAV1 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the B3 DNA Binding Domain of the Arabidopsis Cold-Responsive Transcription Factor RAV1
Plant Cell, 16, 2004
|
|
4ROW
 
 | | The crystal structure of novel APOBEC3G CD2 head-to-tail dimer suggests the binding mode of full-length APOBEC3G to HIV-1 ssDNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Lu, X, Zhang, T, Xu, Z, Liu, S, Zhao, B, Lan, W, Wang, C, Ding, J, Cao, C. | | Deposit date: | 2014-10-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA
J.Biol.Chem., 290, 2015
|
|
4ROV
 
 | | The crystal structure of novel APOBEC3G CD2 head-to-tail dimer suggests the binding mode of full-length APOBEC3G to HIV-1 ssDNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Lu, X, Zhang, T, Xu, Z, Liu, S, Zhao, B, Lan, W, Wang, C, Ding, J, Cao, C. | | Deposit date: | 2014-10-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA
J.Biol.Chem., 290, 2015
|
|
3DFV
 
 | | Adjacent GATA DNA binding | | Descriptor: | DNA (5'-D(*DAP*DAP*DGP*DCP*DAP*DGP*DAP*DTP*DAP*DAP*DGP*DTP*DCP*DTP*DTP*DAP*DTP*DCP*DAP*DG)-3'), DNA (5'-D(*DTP*DTP*DCP*DTP*DGP*DAP*DTP*DAP*DAP*DGP*DAP*DCP*DTP*DTP*DAP*DTP*DCP*DTP*DGP*DC)-3'), Trans-acting T-cell-specific transcription factor GATA-3, ... | | Authors: | Bates, D.L, Kim, G.K, Guo, L, Chen, L. | | Deposit date: | 2008-06-12 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA.
J.Mol.Biol., 381, 2008
|
|
1DUX
 
 | | ELK-1/DNA STRUCTURE REVEALS HOW RESIDUES DISTAL FROM DNA-BINDING SURFACE AFFECT DNA-RECOGNITION | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*T)-3'), ETS-DOMAIN PROTEIN ELK-1 | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 2000-01-19 | | Release date: | 2000-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA.
Nat.Struct.Biol., 7, 2000
|
|
4QR9
 
 | | Crystal structure of two HMGB1 Box A domains cooperating to underwind and kink a DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*GP*AP*TP*AP*T)-3'), High mobility group protein B1, MAGNESIUM ION | | Authors: | Sanchez-Giraldo, R, Acosta-Reyes, F.J, Malarkey, C.S, Saperas, N, Churchill, M.E.A, Campos, J.L. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two high-mobility group box domains act together to underwind and kink DNA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1HCP
 
 | |
8EFC
 
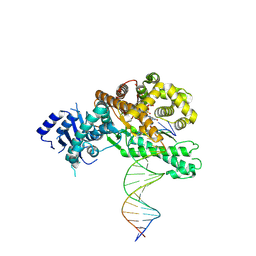 | | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*TP*GP*TP*GP*AP*GP*GP*CP*AP*TP*CP*CP*GP*TP*AP*GP*(2DA))-3'), DNA (5'-D(*AP*GP*CP*TP*CP*TP*AP*CP*GP*GP*AP*TP*GP*CP*CP*TP*CP*AP*CP*AP*G)-3'), ... | | Authors: | Li, C, Zhu, H, Sun, J, Gao, Y. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of DNA polymerase theta mediated DNA end joining.
Nucleic Acids Res., 51, 2023
|
|
3JTG
 
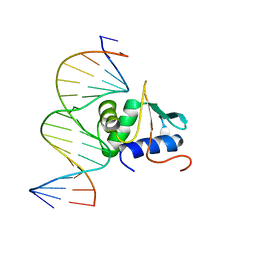 | | Crystal structure of mouse Elf3 C-terminal DNA-binding domain in complex with type II TGF-beta receptor promoter DNA | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*CP*AP*GP*GP*AP*AP*AP*CP*TP*CP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*GP*TP*TP*TP*CP*CP*TP*GP*TP*TP*T)-3'), ETS-related transcription factor Elf-3 | | Authors: | Tahirov, T.H, Babayeva, N.D, Agarkar, V.B, Rizzino, A. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of mouse Elf3 C-terminal DNA-binding domain in complex with type II TGF-beta receptor promoter DNA.
J.Mol.Biol., 397, 2010
|
|
6M2V
 
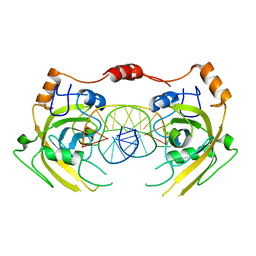 | | Crystal structure of UHRF1 SRA complexed with fully-mCHG DNA. | | Descriptor: | DNA (5'-D(*TP*CP*AP*CP*GP*(5CM)P*TP*GP*CP*GP*TP*GP*A)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Abhishek, S, Nakarakanti, N.K, Deeksha, W, Rajakumara, E. | | Deposit date: | 2020-03-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic insights into recognition of symmetric methylated cytosines in CpG and non-CpG DNA by UHRF1 SRA.
Int.J.Biol.Macromol., 170, 2021
|
|
6M3L
 
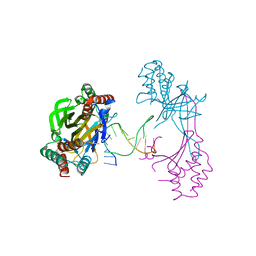 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(nonspecific) complex | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*CP*GP*AP*TP*TP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*GP*CP*G)-3'), RE_R_Pab1 domain-containing protein | | Authors: | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
4REC
 
 | | A nuclease-DNA complex form 3 | | Descriptor: | DNA (40-MER), Fanconi-associated nuclease 1, IODIDE ION | | Authors: | Zhao, Q, Xue, X, Longerich, S, Sung, P, Xiong, Y. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Nat Commun, 5, 2014
|
|
1L3S
 
 | | Crystal Structure of Bacillus DNA Polymerase I Fragment complexed to 9 base pairs of duplex DNA. | | Descriptor: | 5'-D(*GP*A*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*G)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5D5V
 
 | | Crystal structure of human Hsf1 with Satellite III repeat DNA | | Descriptor: | DNA, Heat shock factor protein 1, MAGNESIUM ION | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4LJR
 
 | |
6LMR
 
 | | Solution structure of cold shock domain and ssDNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*CP*T)-3'), Y-box-binding protein 1 | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA binding to human YB-1 cold shock domain regulated by phosphorylation.
Nucleic Acids Res., 48, 2020
|
|
5K58
 
 | |
5CC1
 
 | | S425G Glucocorticoid receptor DNA binding domain - (+)GRE complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*GP*AP*GP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*CP*TP*CP*TP*GP*TP*TP*CP*TP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Hudson, W.H, Weikum, E.A, Ortlund, E.A. | | Deposit date: | 2015-07-01 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Distal substitutions drive divergent DNA specificity among paralogous transcription factors through subdivision of conformational space.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1A6Y
 
 | | REVERBA ORPHAN NUCLEAR RECEPTOR/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*(5IT)P*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*TP*G)-3'), ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | Authors: | Zhao, Q, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 1998-03-04 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elements of an orphan nuclear receptor-DNA complex.
Mol.Cell, 1, 1998
|
|
5JLX
 
 | | AntpHD with 15bp DNA duplex S-monothioated at Cytidine-8 | | Descriptor: | DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*(C7S)P*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), Homeotic protein antennapedia, ... | | Authors: | White, M.A, Zandarashvili, L, Iwahara, J, Nguyen, D. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.748 Å) | | Cite: | Stereospecific Effects of Oxygen-to-Sulfur Substitution in DNA Phosphate on Ion Pair Dynamics and Protein-DNA Affinity.
Chembiochem, 17, 2016
|
|
4LJL
 
 | |
4LJK
 
 | |
