6WJ9
 
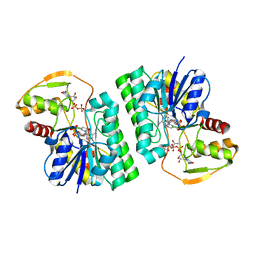 | | UDP-GlcNAc C4-epimerase mutant S121A/Y146F from Pseudomonas protegens in complex with UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Willams, R.J, Whitney, J.C, Whitfield, G.B, Robinson, H, Parsek, M.R, Nitz, M, Harrison, J.J, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
4K3Q
 
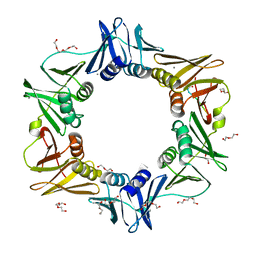 | | E. coli sliding clamp in complex with AcQLDAF | | Descriptor: | (ACE)QLDAF, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
8BNQ
 
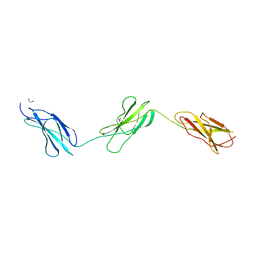 | |
4ZPH
 
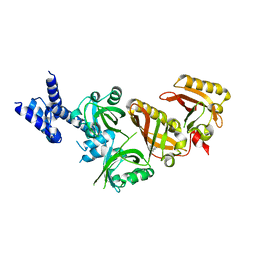 | | Crystal Structure of the Heterodimeric HIF-2a:ARNT Complex with Proflavine | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, PROFLAVIN | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-07 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
6WL0
 
 | | Cryo-EM of Form 1 related peptide filament, 36-31-3-RD | | Descriptor: | peptide 36-31-3-RD | | Authors: | Wang, F, Gnewou, O.M, Su, Z, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
4ZES
 
 | | Blood dendritic cell antigen 2 (BDCA-2) complexed with methyl-mannoside | | Descriptor: | C-type lectin domain family 4 member C, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Jegouzo, S.A.F, Feinberg, H, Dungarwalla, T, Drickamer, K, Weis, W.I, Taylor, M.E. | | Deposit date: | 2015-04-20 | | Release date: | 2015-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Novel Mechanism for Binding of Galactose-terminated Glycans by the C-type Carbohydrate Recognition Domain in Blood Dendritic Cell Antigen 2.
J.Biol.Chem., 290, 2015
|
|
6WV7
 
 | | Human VKOR with Chlorophacinone | | Descriptor: | Chlorophacinone, Vitamin K epoxide reductase, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WVT
 
 | | Structural basis of alphaE-catenin - F-actin catch bond behavior | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Xu, X.P, Pokutta, S, Torres, M, Swift, M.F, Hanein, D, Volkmann, N, Weis, W.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Structural basis of alpha E-catenin-F-actin catch bond behavior.
Elife, 9, 2020
|
|
3QXU
 
 | |
2Z6C
 
 | |
4K7U
 
 | | Crystal structure of Zn2.3-hUb (human ubiquitin) adduct from a solution 70 mM zinc acetate/1.3 mM hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G, Calvaresi, M, Bottoni, A, Arnesano, F, Natile, G. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Conformational selection of ubiquitin quaternary structures driven by zinc ions.
Chemistry, 19, 2013
|
|
8BI3
 
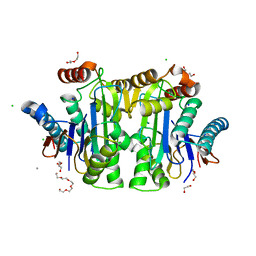 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200W (crystal M200W#1) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-01 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
8BKF
 
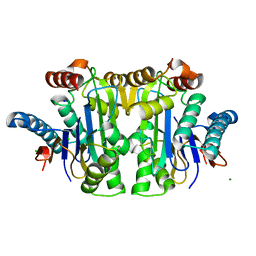 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200T (crystal M200T#o) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase subunit alpha, Isoaspartyl peptidase subunit beta, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-09 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.221 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
8BB6
 
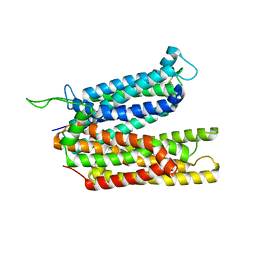 | |
6WVM
 
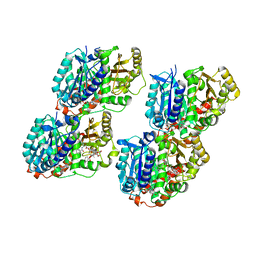 | | High curvature lateral interaction within a 13-protofilament, Taxol stabilized microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Debs, G.E, Cha, M, Huehn, A.R, Sindelar, C.V. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dynamic and asymmetric fluctuations in the microtubule wall captured by high-resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8BP9
 
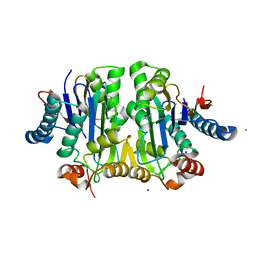 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200W (crystal M200W#2) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase subunit alpha, Isoaspartyl peptidase subunit beta, ... | | Authors: | Sciuk, A, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
5A2V
 
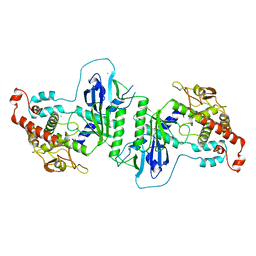 | | Crystal structure of mtPAP in Apo form | | Descriptor: | CHLORIDE ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
8B6U
 
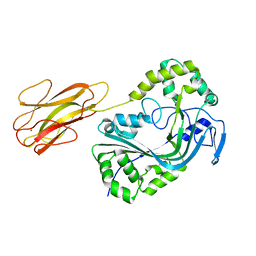 | | Mpf2Ba1 monomer | | Descriptor: | MAGNESIUM ION, Mpf2Ba1 | | Authors: | Marini, G, Poland, B, Leininger, C, Lukoyanova, N, Spielbauer, D, Barry, J, Altier, D, Lum, A, Scolaro, E, Perez-Ortega, C, Yalpani, N, Sandahl, G, Mabry, T, Klever, J, Nowatzki, T, Zhao, J.Z, Sethi, A, Kassa, A, Crane, V, Lu, A, Nelson, M.E, Eswar, N, Topf, M, Saibil, H.R. | | Deposit date: | 2022-09-27 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Structural journey of an insecticidal protein against western corn rootworm.
Nat Commun, 14, 2023
|
|
1EKF
 
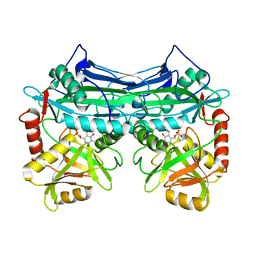 | | CRYSTALLOGRAPHIC STRUCTURE OF HUMAN BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL) COMPLEXED WITH PYRIDOXAL-5'-PHOSPHATE AT 1.95 ANGSTROMS (ORTHORHOMBIC FORM) | | Descriptor: | BRANCHED CHAIN AMINO ACID AMINOTRANSFERASE (MITOCHONDRIAL), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yennawar, N.H, Dunbar, J.H, Conway, M, Hutson, S.M, Farber, G.K. | | Deposit date: | 2000-03-08 | | Release date: | 2001-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5A30
 
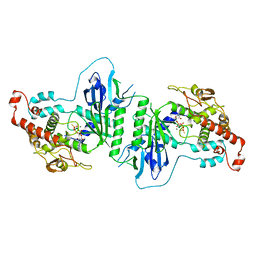 | | Crystal structure of mtPAP N472D mutant in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, MITOCHONDRIAL PROTEIN, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
8B6W
 
 | | Mpf2Ba1 pore | | Descriptor: | MAGNESIUM ION, Mpf2Ba1 | | Authors: | Marini, G, Poland, B, Leininger, C, Lukoyanova, N, Spielbauer, D, Barry, J, Altier, D, Lum, A, Scolaro, E, Perez Ortega, C, Yalpani, N, Sandahl, G, Mabry, T, Klever, J, Nowatzki, T, Zhao, J.Z, Sethi, A, Kassa, A, Crane, V, Lu, A, Nelson, M.E, Eswar, N, Topf, M, Saibil, H.R. | | Deposit date: | 2022-09-27 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural journey of an insecticidal protein against western corn rootworm.
Nat Commun, 14, 2023
|
|
4ZE8
 
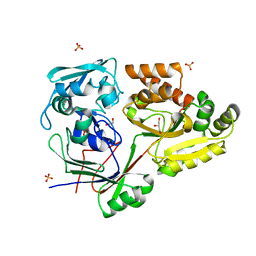 | | PBP AccA from A. tumefaciens C58 | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, substrate binding protein (Agrocinopines A and B), ... | | Authors: | El Sahili, A, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
6WEV
 
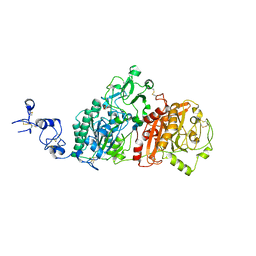 | | Crystal structures of human E-NPP 1: bound to N-{[1-(6,7-dimethoxy-5,8-dihydroquinazolin-4-yl)piperidin-4-yl]methyl}sulfuric diamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Peat, T.S, Dennis, M, Newman, J. | | Deposit date: | 2020-04-03 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human ENPP1 in apo and bound forms.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5A2Z
 
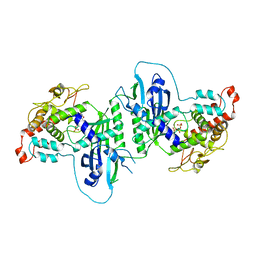 | | Crystal structure of mtPAP in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MITOCHONDRIAL PROTEIN | | Authors: | Lapkouski, M, Hallberg, B.M. | | Deposit date: | 2015-05-26 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of Mitochondrial Poly(A) RNA Polymerase Reveals the Structural Basis for Dimerization, ATP Selectivity and the Spax4 Disease Phenotype.
Nucleic Acids Res., 43, 2015
|
|
8B9Y
 
 | | Cysteine Synthase from Trypanosoma cruzi with PLP and OAS | | Descriptor: | Cysteine synthase, putative, GLYCEROL, ... | | Authors: | Sowerby, K.V, Pohl, E. | | Deposit date: | 2022-10-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cysteine synthase: multiple structures of a key enzyme in cysteine synthesis and a potential drug target for Chagas disease and leishmaniasis.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
