7XB5
 
 | |
7X5H
 
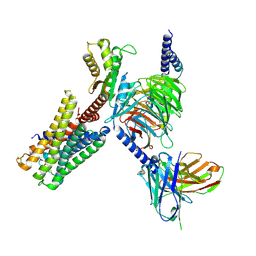 | | Serotonin 5A (5-HT5A) receptor-Gi protein complex | | Descriptor: | 3-(2-azanylethyl)-1H-indole-5-carboxamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tan, Y, Xu, P, Huang, S, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-03-04 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the ligand binding and G i coupling of serotonin receptor 5-HT 5A .
Cell Discov, 8, 2022
|
|
7XHX
 
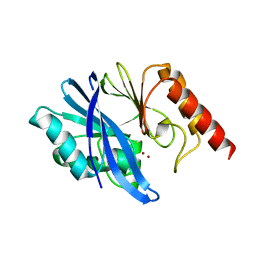 | | Crystal structure of metallo-beta-lactamase IMP-6 | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Yamamoto, K, Tanaka, H, Kurisu, G, Nakano, R, Yano, H, Sakai, H. | | Deposit date: | 2022-04-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the substrate specificity of IMP-6 and IMP-1 metallo-beta-lactamases.
J.Biochem., 173, 2022
|
|
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7XHW
 
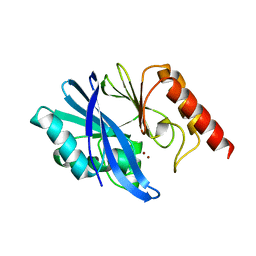 | | Crystal structure of metallo-beta-lactamase IMP-1 | | Descriptor: | Beta-lactamase, ZINC ION | | Authors: | Yamamoto, K, Tanaka, H, Kurisu, G, Nakano, R, Yano, H, Sakai, H. | | Deposit date: | 2022-04-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural insights into the substrate specificity of IMP-6 and IMP-1 metallo-beta-lactamases.
J.Biochem., 173, 2022
|
|
7X4X
 
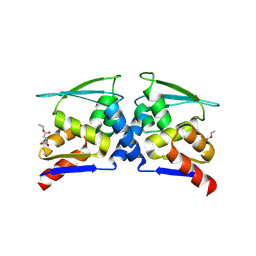 | | BTB domain of KEAP1 in complex with MEF | | Descriptor: | 4-ethoxy-4-oxobutanoic acid, Kelch-like ECH-associated protein 1 | | Authors: | Qu, L.Z. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Characterization of the modification of Kelch-like ECH-associated protein 1 by different fumarates.
Biochem.Biophys.Res.Commun., 605, 2022
|
|
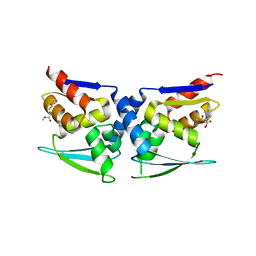
7X4W
 
 | |
7WZZ
 
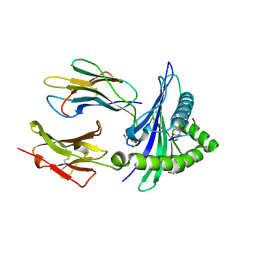 | |
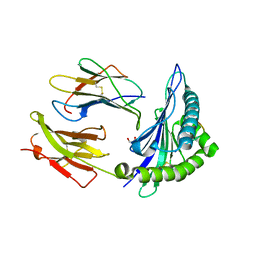
7X00
 
 | |
7X1B
 
 | |
7X1C
 
 | |
7XPN
 
 | |
7XQE
 
 | | Crystal Structure of human RORgamma (C455E) LBD in complex with compound XY039 | | Descriptor: | 2,4-difluoro-N-(1-((4-(trifluoromethyl)benzyl)sulfonyl)-1,2,3,4-tetrahydroquinolin-7-yl)benzenesulfonamide, ETHANOL, GLYCEROL, ... | | Authors: | Wu, X, Li, C, Zhang, Y, Xu, Y. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery and pharmacological characterization of 1,2,3,4-tetrahydroquinoline derivatives as ROR gamma inverse agonists against prostate cancer.
Acta Pharmacol.Sin., 2024
|
|
7XIM
 
 | |
7VM5
 
 | |
7VM6
 
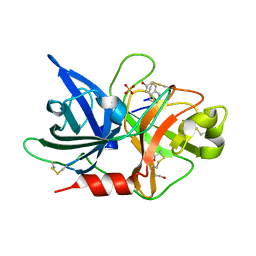 | | Crystal structure of uPA in complex with 6-amidino-2-naphthol | | Descriptor: | 6-oxidanylnaphthalene-2-carboximidamide, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
7VKE
 
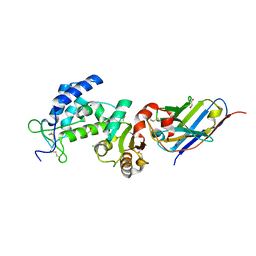 | | Crystal structure of human CD38 ECD in complex with UniDab(TM) F11A | | Descriptor: | 1,2-ETHANEDIOL, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, CHLORIDE ION, ... | | Authors: | Schooten, W.V, Schellenberger, U, Ugamraj, H.S, Manicka, S, Bijpuria, S, Gondu, R.K. | | Deposit date: | 2021-09-29 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TNB-738, a biparatopic antibody, boosts intracellular NAD+ by inhibiting CD38 ecto-enzyme activity.
Mabs, 14, 2022
|
|
7VLI
 
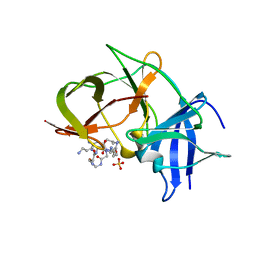 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2220 | | Descriptor: | 1-[(3~{S},6~{S},19~{R})-3,6-bis(4-azanylbutyl)-2,5,8,12,15,18-hexakis(oxidanylidene)-1,4,7,11,14,17-hexazacyclotricos-19-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7VOA
 
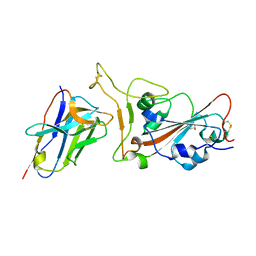 | | Crystal structure of SARS-CoV-2 RBD in complex with aRBD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpaca nanobody | | Authors: | Ma, H, Zeng, W.H, Jin, T.C. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hetero-bivalent nanobodies provide broad-spectrum protection against SARS-CoV-2 variants of concern including Omicron.
Cell Res., 32, 2022
|
|
7VLG
 
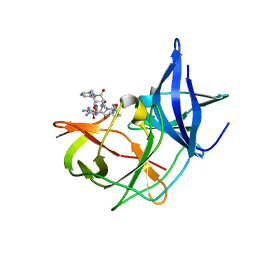 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2201 | | Descriptor: | 1-[(5~{R},8~{R},15~{S},18~{S})-15,18-bis(4-azanylbutyl)-4,7,14,17,20-pentakis(oxidanylidene)-5-propan-2-yl-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7VLH
 
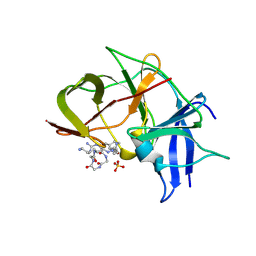 | | Crystal structure of Zika NS2B-NS3 protease with compound MI2219 | | Descriptor: | 1-[(3~{S},6~{R},18~{R})-3,6-bis(4-azanylbutyl)-2,5,8,11,14,17-hexakis(oxidanylidene)-1,4,7,10,13,16-hexazacyclodocos-18-yl]guanidine, NS3 protease, SULFATE ION, ... | | Authors: | Quek, J.P. | | Deposit date: | 2021-10-02 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7Y7U
 
 | |
7W4M
 
 | | Deactive state CI from Q1-NADH dataset, Subclass 4 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W4L
 
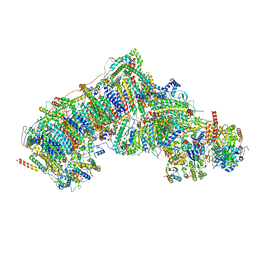 | | Deactive state CI from Q1-NADH dataset, Subclass 3 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W4G
 
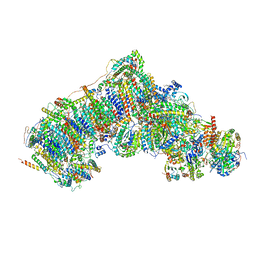 | | Active state CI from Q1-NADH dataset, Subclass 5 | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Gu, J, Yang, M. | | Deposit date: | 2021-11-27 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian mitochondrial complex I.
Nat.Struct.Mol.Biol., 29, 2022
|
|
