1NDV
 
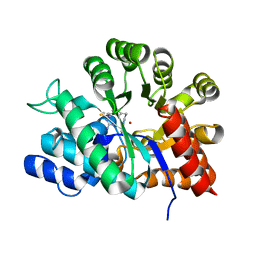 | | Crystal Structure of Adenosine Deaminase complexed with FR117016 | | Descriptor: | Adenosine deaminase, N''-(4-(5-((1H-BENZIMIDAZOL-2-YLAMINO)METHYL)-2-THIENYL)-1,3-THIAZOL-2-YL)GUANIDINE, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2002-12-09 | | Release date: | 2003-12-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A highly potent non-nucleoside adenosine deaminase inhibitor: efficient drug discovery by intentional lead hybridization
J.Am.Chem.Soc., 126, 2004
|
|
1IDG
 
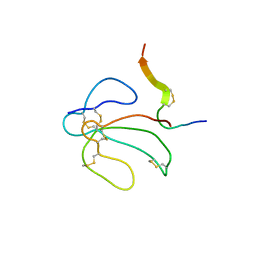 | | THE NMR SOLUTION STRUCTURE OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND AN 18MER COGNATE PEPTIDE | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN, ALPHA-BUNGAROTOXIN | | Authors: | Zeng, H, Moise, L, Grant, M.A, Hawrot, E. | | Deposit date: | 2001-04-04 | | Release date: | 2001-04-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex formed between alpha-bungarotoxin and an 18-mer cognate peptide derived from the alpha 1 subunit of the nicotinic acetylcholine receptor from Torpedo californica.
J.Biol.Chem., 276, 2001
|
|
7QXM
 
 | |
1KSG
 
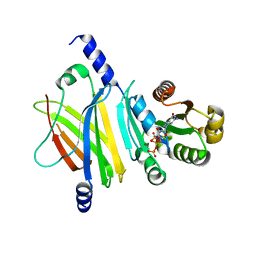 | | Complex of Arl2 and PDE delta, Crystal Form 1 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RETINAL ROD RHODOPSIN-SENSITIVE CGMP 3',5'-CYCLIC PHOSPHODIESTERASE DELTA-SUBUNIT, ... | | Authors: | Hanzal-Bayer, M, Renault, L, Roversi, P, Wittinghofer, A, Hillig, R.C. | | Deposit date: | 2002-01-13 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The complex of Arl2-GTP and PDE delta: from structure to function.
EMBO J., 21, 2002
|
|
1KU6
 
 | | Fasciculin 2-Mouse Acetylcholinesterase Complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Burmeister, W, Taylor, P, Marchot, P. | | Deposit date: | 2002-01-21 | | Release date: | 2003-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site.
EMBO J., 22, 2003
|
|
1KTM
 
 | |
7RM8
 
 | |
1L2X
 
 | | Atomic Resolution Crystal Structure of a Viral RNA Pseudoknot | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA pseudoknot, ... | | Authors: | Egli, M, Minasov, G, Su, L, Rich, A. | | Deposit date: | 2002-02-25 | | Release date: | 2002-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Metal ions and flexibility in a viral RNA pseudoknot at atomic resolution.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8AG6
 
 | |
1LFD
 
 | | CRYSTAL STRUCTURE OF THE ACTIVE RAS PROTEIN COMPLEXED WITH THE RAS-INTERACTING DOMAIN OF RALGDS | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RALGDS, ... | | Authors: | Huang, L, Hofer, F, Martin, G.S, Kim, S.-H. | | Deposit date: | 1998-04-29 | | Release date: | 1999-05-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the interaction of Ras with RalGDS.
Nat.Struct.Biol., 5, 1998
|
|
1LD7
 
 | | Co-crystal structure of Human Farnesyltransferase with farnesyldiphosphate and inhibitor compound 66 | | Descriptor: | (20S)-19,20,22,23-TETRAHYDRO-19-OXO-5H,21H-18,20-ETHANO-12,14-ETHENO-6,10-METHENOBENZ[D]IMIDAZO[4,3-L][1,6,9,13]OXATRIA ZACYCLONOADECOSINE-9-CARBONITRILE, FARNESYL DIPHOSPHATE, ZINC ION, ... | | Authors: | Taylor, J.S, Terry, K.L, Beese, L.S. | | Deposit date: | 2002-04-08 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-Aminopyrrolidinone farnesyltransferase inhibitors: design of macrocyclic compounds with improved pharmacokinetics and excellent cell potency.
J.Med.Chem., 45, 2002
|
|
1L3D
 
 | |
1L1N
 
 | | POLIOVIRUS 3C PROTEINASE | | Descriptor: | Genome polyprotein: Picornain 3C | | Authors: | Mosimann, S.C, Chernaia, M.M, Sia, S, Plotch, S, James, M.N.G. | | Deposit date: | 2002-02-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined X-ray crystallographic structure of the poliovirus 3C gene product.
J.Mol.Biol., 273, 1997
|
|
1L4W
 
 | | NMR structure of an AChR-peptide (Torpedo Californica, alpha-subunit residues 182-202) in complex with alpha-Bungarotoxin | | Descriptor: | Acetylcholine receptor protein, alpha-Bungarotoxin | | Authors: | Samson, A.O, Scherf, T, Rodriguez, E, Eisenstein, M, Anglister, J. | | Deposit date: | 2002-03-06 | | Release date: | 2002-07-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The mechanism for acetylcholine receptor inhibition by alpha-neurotoxins and species-specific resistance to alpha-bungarotoxin revealed by NMR.
Neuron, 35, 2002
|
|
1HH4
 
 | | Rac1-RhoGDI complex involved in NADPH oxidase activation | | Descriptor: | GERAN-8-YL GERAN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Grizot, S, Faure, J, Fieschi, F, Vignais, P.V, Dagher, M.-C, Pebay-Peyroula, E. | | Deposit date: | 2000-12-20 | | Release date: | 2001-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Rac1-Rhogdi Complex Involved in Nadph Oxidase Activation
Biochemistry, 40, 2001
|
|
7R6K
 
 | | State E2 nucleolar 60S ribosomal intermediate - Model for Noc2/Noc3 region | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-22 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R7C
 
 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - L1 stalk local model | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-24 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R72
 
 | | State E1 nucleolar 60S ribosome biogenesis intermediate - Spb4 local model | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-24 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R6Q
 
 | | State E2 nucleolar 60S ribosome biogenesis intermediate - Foot region model | | Descriptor: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-23 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7R7A
 
 | | State E1 nucleolar 60S ribosome biogenesis intermediate - Composite model | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-24 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1MP8
 
 | | Crystal structure of Focal Adhesion Kinase (FAK) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, focal adhesion kinase 1 | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C.G, Pavletich, N.P, Rodgers, J, Sang, B.-C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-11 | | Release date: | 2003-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography
Structure, 10, 2002
|
|
8AEV
 
 | | Human acetylcholinesterase in complex with N,N,N-trimethyl-2-oxo-2-(2-(pyridin-2-ylmethylene)hydrazineyl)ethan-1-aminium | | Descriptor: | 1-(2-(2-((6-(dihydroxymethyl)-2-phenylpyrimidin-4-yl)methylene)hydrazineyl)-2-oxoethyl)pyridin-1-ium, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nachon, F, Dias, J, Brazzolotto, X. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Grid-Type Quaternary Metallosupramolecular Compounds Inhibit Human Cholinesterases through Dynamic Multivalent Interactions.
Chembiochem, 23, 2022
|
|
8AEN
 
 | | Human acetylcholinesterase in complex with zinc and N,N,N-trimethyl-2-oxo-2-(2-(pyridin-2-ylmethylene)hydrazineyl)ethan-1-aminium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Nachon, F, Dias, J, Brazzolotto, X. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Grid-Type Quaternary Metallosupramolecular Compounds Inhibit Human Cholinesterases through Dynamic Multivalent Interactions.
Chembiochem, 23, 2022
|
|
7RDG
 
 | |
1GZM
 
 | | Structure of Bovine Rhodopsin in a Trigonal Crystal Form | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Li, J. | | Deposit date: | 2002-05-24 | | Release date: | 2003-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of Bovine Rhodopsin in a Trigonal Crystal Form
J.Mol.Biol., 343, 2004
|
|
