1V6K
 
 | | Peanut lectin-lactose complex in the presence of peptide(IWSSAGNVA) | | Descriptor: | CALCIUM ION, Galactose-binding lectin, MANGANESE (II) ION, ... | | Authors: | Kundhavai Natchiar, S, Arockia Jeyaprakash, A, Ramya, T.N.C, Thomas, C.J, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-12-01 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural plasticity of peanut lectin: an X-ray analysis involving variation in pH, ligand binding and crystal structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1KVF
 
 | |
4FBU
 
 | | Dpo4 polymerase pre-insertion binary complex with the N-(deoxyguanosin-8-yl)-1-aminopyrene lesion | | Descriptor: | CALCIUM ION, DNA polymerase IV, DNA primer, ... | | Authors: | Kirouac, K, Basu, A, Ling, H. | | Deposit date: | 2012-05-23 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism of replication stalling on a bulky amino-polycyclic aromatic hydrocarbon DNA adduct by a y family DNA polymerase.
J.Mol.Biol., 425, 2013
|
|
4CWC
 
 | |
4PF7
 
 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | (2S)-2-amino-N-{(1S)-1-cyclohexyl-2-[(4-methylphenyl)amino]-2-oxoethyl}-4-(methylselanyl)butanamide, Insulin-degrading enzyme, ZINC ION | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
1VZM
 
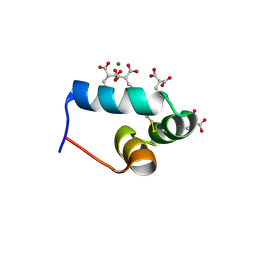 | | OSTEOCALCIN FROM FISH ARGYROSOMUS REGIUS | | Descriptor: | MAGNESIUM ION, OSTEOCALCIN | | Authors: | Frazao, C, Simes, D.C, Coelho, R, Alves, D, Williamson, M.K, Price, P.A, Cancela, M.L, Carrondo, M.A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-09-10 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Evidence of a Fourth Gla Residue in Fish Osteocalcin: Biological Implications
Biochemistry, 44, 2005
|
|
4BCG
 
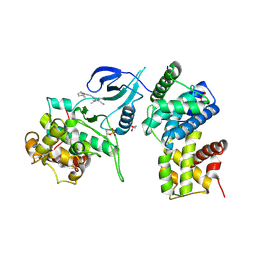 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 2-[[3-(1,4-diazepan-1-yl)phenyl]amino]-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidine-5-carbonitrile, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1, ... | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.085 Å) | | Cite: | Substituted 4-(Thiazol-5-Yl)-2-(Phenylamino)Pyrimidines are Highly Active Cdk9 Inhibitors: Synthesis, X-Ray Crystal Structure, Sar and Anti-Cancer Activities.
J.Med.Chem., 56, 2013
|
|
1PV8
 
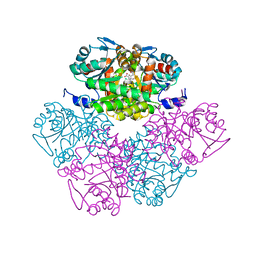 | | Crystal structure of a low activity F12L mutant of human porphobilinogen synthase | | Descriptor: | 3-(2-AMINOETHYL)-4-(AMINOMETHYL)HEPTANEDIOIC ACID, Delta-aminolevulinic acid dehydratase, ZINC ION | | Authors: | Breinig, S, Kervinen, J, Stith, L, Wasson, A.S, Fairman, R, Wlodawer, A, Zdanov, A, Jaffe, E.K. | | Deposit date: | 2003-06-26 | | Release date: | 2003-09-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase.
Nat.Struct.Biol., 10, 2003
|
|
1PJL
 
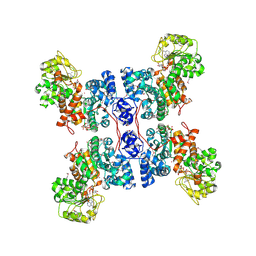 | | Crystal structure of human m-NAD-ME in ternary complex with NAD and Lu3+ | | Descriptor: | LUTETIUM (III) ION, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Yang, Z, Batra, R, Floyd, D.L, Hung, H.-C, Chang, G.-G, Tong, L. | | Deposit date: | 2003-06-03 | | Release date: | 2003-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potent and competitive inhibition of malic enzymes by lanthanide ions
Biochem.Biophys.Res.Commun., 274, 2000
|
|
4RFE
 
 | | Crystal structure of ADCC-potent ANTI-HIV-1 Rhesus macaque antibody JR4 Fab | | Descriptor: | CHLORIDE ION, Fab heavy chain of ADCC-potent anti-HIV-1 antibody JR4, Fab light chain of ADCC-potent anti-HIV-1 antibody JR4, ... | | Authors: | Wu, X, Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
4JHL
 
 | | Crystal Structure of of Axe2, an Acetylxylan Esterase from Geobacillus stearothermophilus | | Descriptor: | Acetyl xylan esterase, CHLORIDE ION, GLYCEROL | | Authors: | Lansky, S, Alalouf, O, Solomon, V, Alhassid, A, Belrahli, H, Govada, L, Chayan, N.E, Shoham, Y, Shoham, G. | | Deposit date: | 2013-03-05 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A unique octameric structure of Axe2, an intracellular acetyl-xylooligosaccharide esterase from Geobacillus stearothermophilus.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1DO2
 
 | | TRIGONAL CRYSTAL FORM OF HEAT SHOCK LOCUS U (HSLU) FROM ESCHERICHIA COLI | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN (HEAT SHOCK LOCUS U) | | Authors: | Bochtler, M, Hartmann, C, Song, H.K, Bourenkov, G.P, Bartunik, H.D. | | Deposit date: | 1999-12-18 | | Release date: | 2000-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The structures of HsIU and the ATP-dependent protease HsIU-HsIV.
Nature, 403, 2000
|
|
4S2E
 
 | |
4FN7
 
 | | Apo Structure of the Mtb enoyol CoA isomerase (Rv0632c) | | Descriptor: | CHLORIDE ION, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Bruning, J.B, Gao, N, Hernandez, E.D, Li, H, Dang, N, Hung, L.W, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-06-19 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | "Crystal Structure and Mechanism of the Prokaryotic Enoyl-CoA Isomerase (ECI)"
To be Published
|
|
4FOL
 
 | | S-formylglutathione hydrolase Variant H160I | | Descriptor: | S-formylglutathione hydrolase | | Authors: | Legler, P.M, Millard, C.B. | | Deposit date: | 2012-06-20 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A role for His-160 in peroxide inhibition of S. cerevisiae S-formylglutathione hydrolase: Evidence for an oxidation sensitive motif.
Arch.Biochem.Biophys., 528, 2012
|
|
4FN8
 
 | | Crystal structure of the Mtb enoyl CoA isomerase (Rv0632c)in complex with acetoacetyl CoA | | Descriptor: | ACETOACETYL-COENZYME A, Enoyl-CoA hydratase/isomerase family protein, SULFATE ION | | Authors: | Bruning, J.B, Gao, N, Hernandez, E.D, Li, H, Dang, N, Hung, L.W, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-06-19 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | Crystal structure and mechanism of the prokaryotic enoyl CoA isomerase (ECI)
To be Published
|
|
4FNB
 
 | | Crystal structure of the Mtb enoyl CoA isomerase (Rv0632c) in complex with hydroxybutyrl CoA | | Descriptor: | 3-HYDROXYBUTANOYL-COENZYME A, Enoyl-CoA hydratase/isomerase family protein, SULFATE ION | | Authors: | Bruning, J.B, Gao, N, Hernandez, E.D, Li, H, Dang, N, Hung, L.W, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-06-19 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Mechanism of the Prokaryotic Enoyl CoA Isomerase
To be Published
|
|
1NBM
 
 | | THE STRUCTURE OF BOVINE F1-ATPASE COVALENTLY INHIBITED WITH 4-CHLORO-7-NITROBENZOFURAZAN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, F1-ATPASE, ... | | Authors: | Orriss, G.L, Leslie, A.G.W, Braig, K, Walker, J.E. | | Deposit date: | 1998-04-30 | | Release date: | 1998-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bovine F1-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: the structure provides further support for a rotary catalytic mechanism.
Structure, 6, 1998
|
|
2XC0
 
 | | Factor Xa in complex with a pyrrolidine-3,4-dicarboxylic acid inhibitor | | Descriptor: | (3R,4R)-1-METHANESULFONYL-PYRROLIDINE-3,4--DICARBOXYLIC ACID 3-[(3-FLUORO-4-METHOXY-PHENYL)-AMIDE] 4-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYL]-AMIDE}, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Banner, D.W, Benz, J, Schlatter, D, Anselm, L, Haap, W. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a Factor Xa Inhibitor (3R,4R)-1-(2,2-Difluoro-Ethyl)-Pyrrolidine-3,4-Dicarboxylic Acid 3-[(5-Chloro-Pyridin-2-Yl)-Amide] 4-{[2-Fluoro-4-(2-Oxo-2H-Pyridin-1-Yl)-Phenyl]-Amide} as a Clinical Candidate.
Bioorg.Med.Chem., 20, 2010
|
|
4TPM
 
 | |
4JAQ
 
 | | Crystal Structure of Mycobacterium tuberculosis PKS11 Reveals Intermediates in the Synthesis of Methyl-branched Alkylpyrones | | Descriptor: | 3,5-dioxoicosanoic acid, Alpha-pyrone synthesis polyketide synthase-like Pks11, COENZYME A, ... | | Authors: | Gokulan, K, Sacchettini, J.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2013-02-19 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis polyketide synthase 11 (PKS11) reveals intermediates in the synthesis of methyl-branched alkylpyrones.
J.Biol.Chem., 288, 2013
|
|
4JE6
 
 | | Crystal structure of a human-like mitochondrial peptide deformylase | | Descriptor: | Peptide deformylase 1A, chloroplastic/mitochondrial, ZINC ION | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JD3
 
 | |
4DUX
 
 | | E. coli (lacZ) beta-galactosidase (N460S) in complex with L-ribose | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Wheatley, R.W, Lo, S, Janzcewicz, L.J, Dugdale, M.L, Huber, R.E. | | Deposit date: | 2012-02-22 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural explanation for allolactose (lac operon inducer) synthesis by lacZ beta-galactosidase and the evolutionary relationship between allolactose synthesis and the lac repressor.
J.Biol.Chem., 288, 2013
|
|
4JY1
 
 | | CRYSTAL STRUCTURE OF HCV NS5B POLYMERASE IN COMPLEX WITH COMPOUND 5 | | Descriptor: | 3-{ISOPROPYL[(TRANS-4-METHYLCYCLOHEXYL)CARBONYL]AMINO}-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, Genome polyprotein | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinguishing drug binding pockets on proteins by complementary biophysical and biological methods
To be Published
|
|
