1VHG
 
 | | Crystal structure of ADP compounds hydrolase | | Descriptor: | ADP compounds hydrolase nudE | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHT
 
 | |
1VI9
 
 | | Crystal structure of pyridoxamine kinase | | Descriptor: | BETA-MERCAPTOETHANOL, Pyridoxamine kinase, SULFATE ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VH2
 
 | |
1VH9
 
 | | Crystal structure of a putative thioesterase | | Descriptor: | Hypothetical protein ybdB | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
7A4Q
 
 | | The Crystal structure of RO4613269 bound to CK2alpha | | Descriptor: | 2-methoxyimino-5-(quinolin-6-ylmethyl)-1,3-thiazol-4-one, Casein kinase II subunit alpha | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2020-08-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Chemical proteomics reveals the target landscape of 1,000 kinase inhibitors.
Nat.Chem.Biol., 2023
|
|
6KTX
 
 | | The wildtype structure of EanB | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
2MFR
 
 | |
2QMV
 
 | |
2MRU
 
 | | Structure of truncated EcMazE-DNA complex | | Descriptor: | Antitoxin MazE, DNA (5'-D(*CP*GP*TP*GP*AP*TP*AP*TP*AP*TP*AP*GP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*CP*TP*AP*TP*AP*TP*AP*TP*CP*AP*CP*G)-3') | | Authors: | Zorzini, V, Buts, L, Loris, R, van Nuland, N. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Escherichia coli antitoxin MazE as transcription factor: insights into MazE-DNA binding.
Nucleic Acids Res., 43, 2015
|
|
7MLP
 
 | | Crystal structure of ricin A chain in complex with 5-(2,6-dimethylphenyl)thiophene-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-(2,6-dimethylphenyl)thiophene-2-carboxylic acid, Ricin, ... | | Authors: | Harijan, R.K, Li, X.P, Cao, B, Augeri, D, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2021-04-28 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Synthesis and Structural Characterization of Ricin Inhibitors Targeting Ribosome Binding Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7MLN
 
 | | Crystal structure of ricin A chain in complex with 5-(o-tolyl)thiophene-2-carboxylic acid | | Descriptor: | 5-(2-methylphenyl)thiophene-2-carboxylic acid, GLYCEROL, Ricin | | Authors: | Harijan, R.K, Li, X.P, Cao, B, Augeri, D, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2021-04-28 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Synthesis and Structural Characterization of Ricin Inhibitors Targeting Ribosome Binding Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7MLO
 
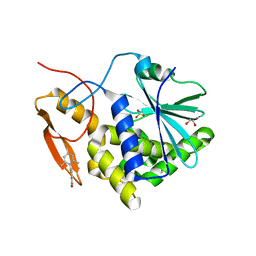 | | Crystal structure of ricin A chain in complex with 5-mesitylthiophene-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-(2,4,6-trimethylphenyl)thiophene-2-carboxylic acid, Ricin, ... | | Authors: | Harijan, R.K, Li, X.P, Cao, B, Augeri, D, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2021-04-28 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthesis and Structural Characterization of Ricin Inhibitors Targeting Ribosome Binding Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
7MLT
 
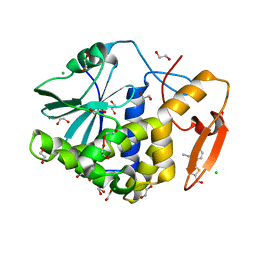 | | Crystal structure of ricin A chain in complex with 5-(2-ethylphenyl)thiophene-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-ethylphenyl)thiophene-2-carboxylic acid, CHLORIDE ION, ... | | Authors: | Harijan, R.K, Li, X.P, Cao, B, Augeri, D, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | Deposit date: | 2021-04-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis and Structural Characterization of Ricin Inhibitors Targeting Ribosome Binding Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
6X19
 
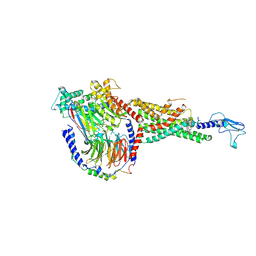 | | Non peptide agonist CHU-128, bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | 3-{(1S)-1-[5-(2,2-dimethylmorpholin-4-yl)-2-{(4S)-2-(4-fluoro-3,5-dimethylphenyl)-3-[3-(4-fluoro-1-methyl-1H-indazol-5-yl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]-4-methyl-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carbonyl}-1H-indol-1-yl]butyl}-1,2,4-oxadiazol-5(4H)-one, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
6X18
 
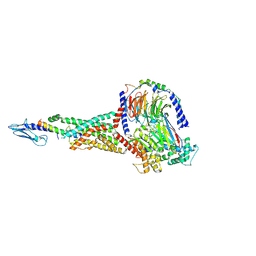 | | GLP-1 peptide hormone bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | Glucagon, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
7NHZ
 
 | |
6R8E
 
 | | SC14 G-hairpin | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*GP*T)-3') | | Authors: | Lenarcic Zivkovic, M, Trantirek, L, Plavec, J. | | Deposit date: | 2019-04-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insight into formation propensity of pseudocircular DNA G-hairpins.
Nucleic Acids Res., 49, 2021
|
|
6R2X
 
 | | NMR structure of Chromogranin A (F39-D63) | | Descriptor: | Chromogranin-A | | Authors: | Nardelli, F, Quilici, G, Ghitti, M, Curnis, F, Gori, A, Berardi, A, Corti, A, Musco, G. | | Deposit date: | 2019-03-19 | | Release date: | 2019-12-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A stapled chromogranin A-derived peptide is a potent dual ligand for integrins alpha v beta 6 and alpha v beta 8.
Chem.Commun.(Camb.), 55, 2019
|
|
2BET
 
 | | Structure of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, in complex with 4-phospho-D-erythronate. | | Descriptor: | 4-PHOSPHO-D-ERYTHRONATE, CARBOHYDRATE-PHOSPHATE ISOMERASE | | Authors: | Roos, A.K, Ericsson, D.J, Mowbray, S.L. | | Deposit date: | 2004-11-30 | | Release date: | 2004-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Competitive Inhibitors of Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase B Reveal New Information About the Reaction Mechanism
J.Biol.Chem., 280, 2005
|
|
2XON
 
 | | Structure of TmCBM61 in complex with beta-1,4-galactotriose at 1.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ARABINOGALACTAN ENDO-1,4-BETA-GALACTOSIDASE, CALCIUM ION, ... | | Authors: | Cid, M, Lodberg-Pedersen, H, Kaneko, S, Coutinho, P.M, Henrissat, B, Willats, W.G.T, Boraston, A.B. | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recognition of the Helical Structure of Beta-1,4-Galactan by a New Family of Carbohydrate-Binding Modules.
J.Biol.Chem., 285, 2010
|
|
2BES
 
 | | Structure of Mycobacterium tuberculosis Ribose-5-Phosphate Isomerase, RpiB, Rv2465c, in complex with 4-phospho-D-erythronohydroxamic acid. | | Descriptor: | 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, CARBOHYDRATE-PHOSPHATE ISOMERASE | | Authors: | Roos, A.K, Ericsson, D.J, Mowbray, S.L. | | Deposit date: | 2004-11-30 | | Release date: | 2004-12-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive Inhibitors of Mycobacterium Tuberculosis Ribose-5-Phosphate Isomerase B Reveal New Information About the Reaction Mechanism.
J.Biol.Chem., 280, 2005
|
|
6YF3
 
 | | FKBP12 in complex with the BMP potentiator compound 10 at 1.00A resolution | | Descriptor: | (1~{R},9~{S},12~{S},13~{R},14~{S},17~{R},18~{E},21~{S},23~{S},24~{R},25~{S},27~{R})-17-ethyl-25-methoxy-12-[(~{E})-1-[(1~{R},3~{R},4~{R})-3-methoxy-4-oxidanyl-cyclohexyl]prop-1-en-2-yl]-13,19,21,27-tetramethyl-1,14,23-tris(oxidanyl)-11,28-dioxa-4-azatricyclo[22.3.1.0^{4,9}]octacos-18-ene-2,3,10,16-tetrone, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Kallen, J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Phenotypic screen identifies calcineurin-sparing FK506 analogs as BMP potentiators for treatment of acute kidney injury.
Cell Chem Biol, 28, 2021
|
|
6Y6R
 
 | | Crystal structure of MINDY1 T335D mutant | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase MINDY-1 | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2020-02-27 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
6YJG
 
 | | Crystal structure of MINDY1 mutant-Y114F | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase MINDY1 | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
