2RV4
 
 | | Solution structures of the DNA-binding domain (ZF5) of mouse immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
8H7T
 
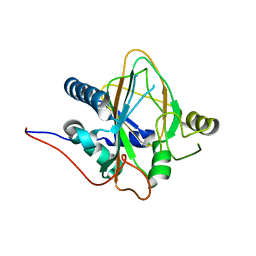 | | Trans-3/4-proline-hydroxylase H11 apo structure | | Descriptor: | CHLORIDE ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
4WAC
 
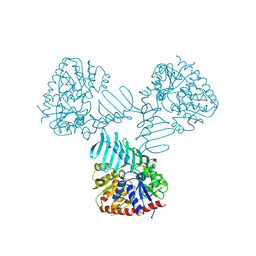 | | Crystal Structure of TarM | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Koc, C, Stehle, T, Xia, G, Peschel, A. | | Deposit date: | 2014-08-29 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Enzymatic Analysis of TarM Glycosyltransferase from Staphylococcus aureus Reveals an Oligomeric Protein Specific for the Glycosylation of Wall Teichoic Acid.
J.Biol.Chem., 290, 2015
|
|
8H00
 
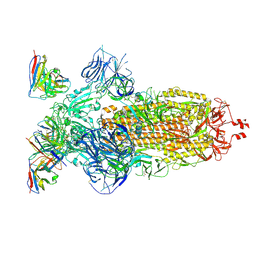 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
2RFK
 
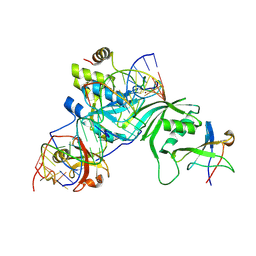 | | Substrate RNA Positioning in the Archaeal H/ACA Ribonucleoprotein Complex | | Descriptor: | Probable tRNA pseudouridine synthase B, Ribosome biogenesis protein Nop10, Small nucleolar rnp similar to gar1, ... | | Authors: | Liang, B, Xue, S, Terns, R.M, Terns, M.P, Li, H, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-09-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8HHM
 
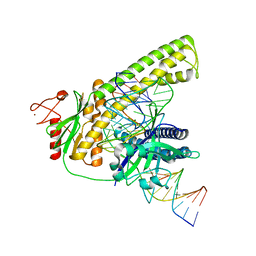 | | Cryo-EM structure of the Cas12m2-crRNA-target DNA ternary complex intermediate state | | Descriptor: | Cas12m2, DNA (36-MER), MAGNESIUM ION, ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2RFV
 
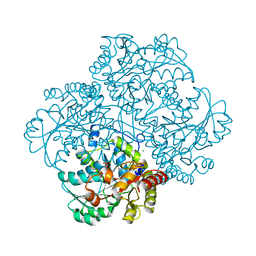 | | High resolution structure of L-methionine gamma-lyase from Citrobacter freundii | | Descriptor: | CHLORIDE ION, Methionine gamma-lyase | | Authors: | Nikulin, A.D, Revtovich, S.V, Morozova, E.A, Nevskaya, N.A, Nikonov, S.V, Garber, M.B, Demidkina, T.V. | | Deposit date: | 2007-10-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.355 Å) | | Cite: | High-resolution structure of methionine gamma-lyase from Citrobacter freundii.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
8HIF
 
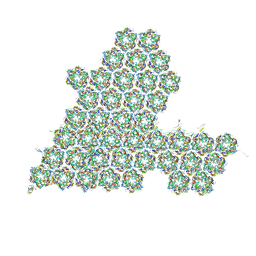 | | One asymmetric unit of Singapore grouper iridovirus capsid | | Descriptor: | Major capsid protein, Penton protein (VP14), VP137, ... | | Authors: | Zhao, Z.N, Liu, C.C, Zhu, D.J, Qi, J.X, Zhang, X.Z, Gao, G.F. | | Deposit date: | 2022-11-20 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic architecture of Singapore grouper iridovirus and implications for giant virus assembly.
Nat Commun, 14, 2023
|
|
2RGA
 
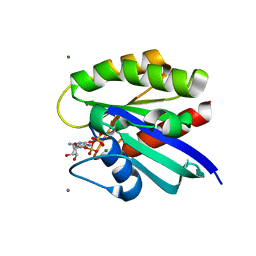 | | Crystal structure of H-RasQ61I-GppNHp | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Buhrman, G, Wink, G, Mattos, C. | | Deposit date: | 2007-10-03 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transformation Efficiency of RasQ61 Mutants Linked to Structural Features of the Switch Regions in the Presence of Raf.
Structure, 15, 2007
|
|
4WB6
 
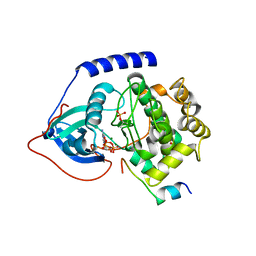 | | Crystal structure of a L205R mutant of human cAMP-dependent protein kinase A (catalytic alpha subunit) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PKI (5-24), ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2SGQ
 
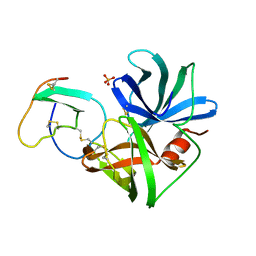 | | GLN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
4WBO
 
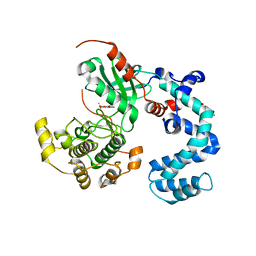 | |
8H81
 
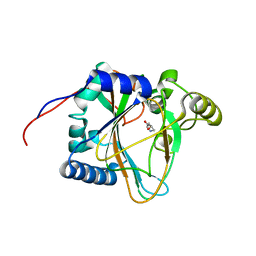 | | Trans-3/4-proline-hydroxylase H11 with 4-Hydroxyl-proline | | Descriptor: | 4-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8HMZ
 
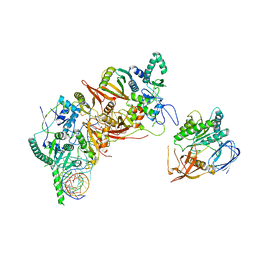 | | Cryo-EM structure of the human post-catalytic TSEN/pre-tRNA complex | | Descriptor: | Chromosome 1 open reading frame 19, isoform CRA_a, MAGNESIUM ION, ... | | Authors: | Zhang, X, Yang, F, Zhan, X, Shi, Y. | | Deposit date: | 2022-12-06 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of pre-tRNA intron removal by human tRNA splicing endonuclease.
Mol.Cell, 83, 2023
|
|
4WD2
 
 | |
2SKE
 
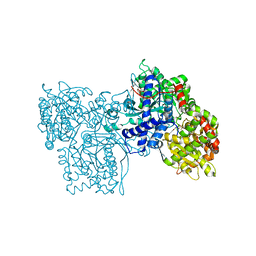 | | PYRIDOXAL PHOSPHORYLASE B IN COMPLEX WITH PHOSPHITE, GLUCOSE AND INOSINE-5'-MONOPHOSPHATE | | Descriptor: | INOSINIC ACID, PHOSPHITE ION, PYRIDOXAL PHOSPHORYLASE B, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Tsitsanou, K.E, Johnson, L.N, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Activator anion binding site in pyridoxal phosphorylase b: the binding of phosphite, phosphate, and fluorophosphate in the crystal.
Protein Sci., 5, 1996
|
|
4WDD
 
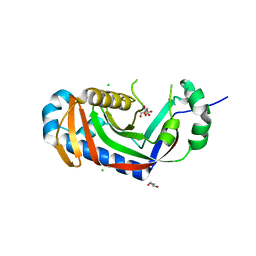 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation T232A, complexed with citrate | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
8HHL
 
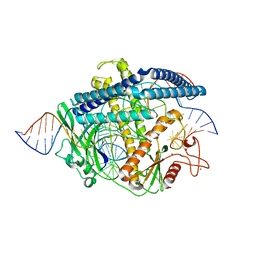 | | Cryo-EM structure of the Cas12m2-crRNA-target DNA full R-loop complex | | Descriptor: | Cas12m2, MAGNESIUM ION, NTS (36-MER), ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2STA
 
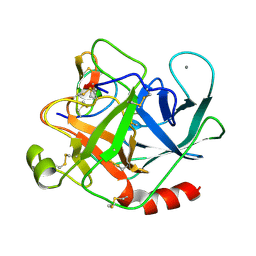 | | ANIONIC SALMON TRYPSIN IN COMPLEX WITH SQUASH SEED INHIBITOR (CUCURBITA MAXIMA TRYPSIN INHIBITOR I) | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Berglund, G.I, Otlewski, J, Apostoluk, W, Andersen, O.A, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-12-10 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of three new trypsin-squash-inhibitor complexes: a detailed comparison with other trypsins and their complexes.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4WDW
 
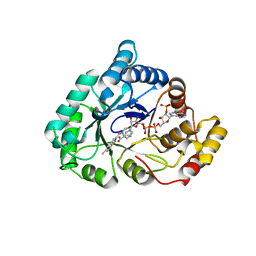 | | 17beta-HSD5 in complex with 3,6-dihydropyridin-1(2H)-yl(5-methyl-1H-indol-2-yl)methanone | | Descriptor: | 3,6-dihydropyridin-1(2H)-yl(5-methyl-1H-indol-2-yl)methanone, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of complexes of type 5 17 beta-hydroxysteroid dehydrogenase with structurally diverse inhibitors: insights into the conformational changes upon inhibitor binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8H17
 
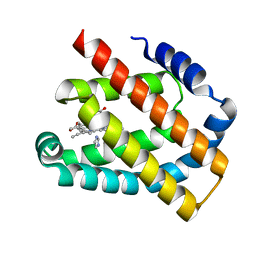 | | Crystal structure of the Globin domain of Thermosynechococcus elongatus BP-1 | | Descriptor: | IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, Tlr1989 protein | | Authors: | Mathur, S, Yadav, S.K, Pal, K.R, Kundu, S. | | Deposit date: | 2022-09-30 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A novel single sensor hemoglobin domain from the thermophilic cyanobacteria Thermosynechococcus elongatus BP-1 exhibits higher pH but lower thermal stability compared to globins from mesophilic organisms.
Int.J.Biol.Macromol., 240, 2023
|
|
2TGI
 
 | |
8HHG
 
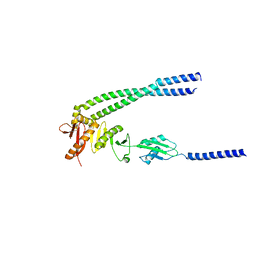 | | The bacterial divisome protein complex FtsB-FtsL-FtsQ | | Descriptor: | Cell division protein FtsB, Cell division protein FtsL, Cell division protein FtsQ | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the heterotrimeric membrane protein complex FtsB-FtsL-FtsQ of the bacterial divisome.
Nat Commun, 14, 2023
|
|
4V91
 
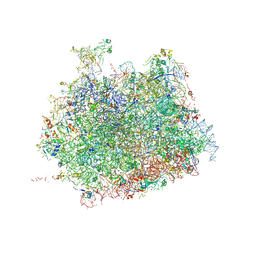 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 25S RRNA, 5.8S RRNA, 5S RRNA, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
8GZG
 
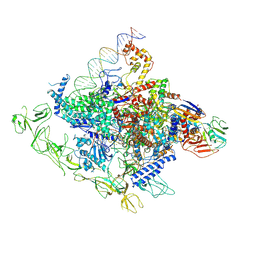 | | Cryo-EM structure of Synechocystis sp. PCC 6803 RPitc | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shen, L.Q, You, L.L, Zhang, Y. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of Synechocystis sp. PCC 6803 CTP-bound RPitc
Proc.Natl.Acad.Sci.USA, 2023
|
|
