5J5E
 
 | | crystal structure of antigen-ERAP1 domain complex | | Descriptor: | Endoplasmic reticulum aminopeptidase 1 | | Authors: | Sui, L, Gandhi, A, Guo, H.-C. | | Deposit date: | 2016-04-02 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a polypeptide's C-terminus in complex with the regulatory domain of ER aminopeptidase 1.
Mol.Immunol., 80, 2016
|
|
5WN5
 
 | | APE1 exonuclease substrate complex with a C/T mismatch and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(DV3))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular snapshots of APE1 proofreading mismatches and removing DNA damage.
Nat Commun, 9, 2018
|
|
9BHK
 
 | | MerTK in complex with small molecule inhibitor 6-{1-[6-(3-hydroxy-3-methylbutoxy)-1,3-benzoxazol-2-yl]azetidin-3-yl}-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide | | Descriptor: | 6-{1-[6-(3-hydroxy-3-methylbutoxy)-1,3-benzoxazol-2-yl]azetidin-3-yl}-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | Jakob, C.G, Gurbani, D, Qiu, W. | | Deposit date: | 2024-04-20 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Discovery of Potent Azetidine-Benzoxazole MerTK Inhibitors with In Vivo Target Engagement.
J.Med.Chem., 67, 2024
|
|
7ZCY
 
 | | Sporosarcina pasteurii urease (SPU) co-crystallized in the presence of an Ebselen-derivative and bound to Se atoms | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, N-(2-chloranyl-4-fluoranyl-phenyl)-2-selanyl-benzamide, ... | | Authors: | Mazzei, L, Ciurli, S, Cianci, M. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-19 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Optimized Ebselen-Based Inhibitors of Bacterial Ureases with Nontypical Mode of Action.
J.Med.Chem., 66, 2023
|
|
7TM4
 
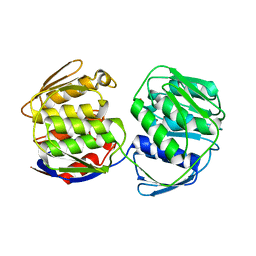 | |
9IW1
 
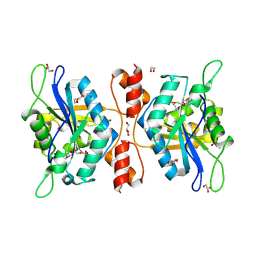 | | wild type NMN/NaMN adenylyltransferase from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Qian, X.-L, Zheng, Y.-C, Chen, C, Xu, J.-H. | | Deposit date: | 2024-07-24 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Biochemical and structural characterization of a novel nicotinamide mononucleotide adenylyltransferase from thermophilic fungi Chaetomium thermophilum.
Biochem.Biophys.Res.Commun., 776, 2025
|
|
9BVH
 
 | | Homomeric alpha3 glycine receptor in the presence of 0.1 mM glycine in an apo state. | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Kindig, K, Gibbs, E, Chakrapani, S. | | Deposit date: | 2024-05-20 | | Release date: | 2024-11-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Mechanisms underlying modulation of human GlyR alpha 3 by Zn 2+ and pH.
Sci Adv, 10, 2024
|
|
9BU3
 
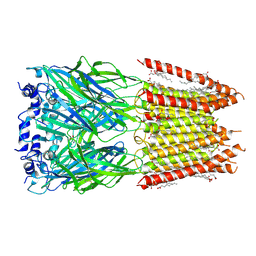 | | Homomeric alpha3 glycine receptor in the presence of 0.1 mM glycine in a desensitized state | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kindig, K, Gibbs, E, Chakrapani, S. | | Deposit date: | 2024-05-16 | | Release date: | 2024-11-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms underlying modulation of human GlyR alpha 3 by Zn 2+ and pH.
Sci Adv, 10, 2024
|
|
7TBG
 
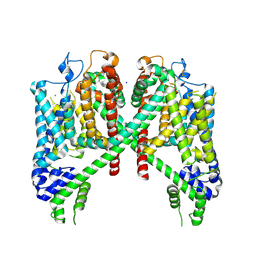 | | AtTPC1 D454N with 1 mM Ca2+ | | Descriptor: | CALCIUM ION, SODIUM ION, Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-22 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7QS8
 
 | | Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 | | Descriptor: | Protein Tax-1, Protein scribble homolog | | Authors: | Javorsky, A, Soares da Costa, T.P, Mackie, E.R, Humbert, P.O, Kvansakul, M. | | Deposit date: | 2022-01-13 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 PBM.
Febs J., 290, 2023
|
|
7TDF
 
 | | AtTPC1 D454N with 1 mM EDTA state I | | Descriptor: | Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9BVJ
 
 | | Homomeric alpha3 glycine receptor in the presence of 1 mM glycine in an desensitized state. | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kindig, K, Gibbs, E, Chakrapani, S. | | Deposit date: | 2024-05-20 | | Release date: | 2024-11-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms underlying modulation of human GlyR alpha 3 by Zn 2+ and pH.
Sci Adv, 10, 2024
|
|
9IO6
 
 | |
7QRT
 
 | | Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 | | Descriptor: | CHLORIDE ION, Protein Tax-1, Protein scribble homolog, ... | | Authors: | Javorsky, A, Soares da Costa, T.P, Mackie, E.R, Humbert, P.O, Kvansakul, M, Maddumage, J.C. | | Deposit date: | 2022-01-12 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into the Scribble PDZ domains interaction with the oncogenic Human T-cell lymphotrophic virus-1 (HTLV-1) Tax1 PBM.
Febs J., 290, 2023
|
|
6GQM
 
 | | Crystal structure of human c-KIT kinase domain in complex with a small molecule inhibitor, AZD3229 | | Descriptor: | CHLORIDE ION, Mast/stem cell growth factor receptor Kit, ~{N}-[4-[[5-fluoranyl-7-(2-methoxyethoxy)quinazolin-4-yl]amino]phenyl]-2-(4-propan-2-yl-1,2,3-triazol-1-yl)ethanamide | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
5MEH
 
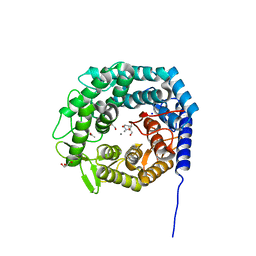 | |
6GPU
 
 | | Crystal structure of miniSOG at 1.17A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Lafaye, C, Signor, L, Aumonier, S, Shu, X, Gotthard, G, Royant, A. | | Deposit date: | 2018-06-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Tailing miniSOG: structural bases of the complex photophysics of a flavin-binding singlet oxygen photosensitizing protein.
Sci Rep, 9, 2019
|
|
6GQK
 
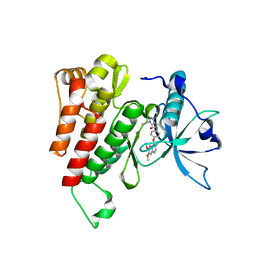 | | Crystal structure of human c-KIT kinase domain in complex with AZD3229-analogue (compound 23) | | Descriptor: | Mast/stem cell growth factor receptor Kit, ~{N}-[4-(6,7-dimethoxyquinazolin-4-yl)oxyphenyl]-2-(1-ethylpyrazol-4-yl)ethanamide | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
7A6J
 
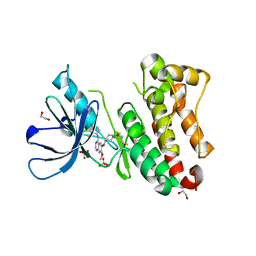 | | Crystal Structure of EGFR-T790M/V948R in Complex with Poziotinib | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-[[3,4-bis(chloranyl)-2-fluoranyl-phenyl]amino]-7-methoxy-quinazolin-6-yl]oxypiperidin-1-yl]propan-1-one, Epidermal growth factor receptor, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2020-08-25 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into Targeting Exon20 Insertion Mutations of the Epidermal Growth Factor Receptor with Wild Type-Sparing Inhibitors.
J.Med.Chem., 65, 2022
|
|
9F2N
 
 | | Structure of human carbonic anhydrase XII complexed with 3-(cyclooctylamino)-2,6-difluoro-4-((3-hydroxypropyl)sulfonyl)-5-(piperidin-1-yl)benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 3-(cyclooctylamino)-2,6-difluoro-4-((3-hydroxypropyl)sulfonyl)-5-(piperidin-1-yl)benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Manakova, E.N, Grazulis, S, Paketuryte, V, Smirnov, A. | | Deposit date: | 2024-04-23 | | Release date: | 2025-05-14 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Di- meta -Substituted Fluorinated Benzenesulfonamides as Potent and Selective Anticancer Inhibitors of Carbonic Anhydrase IX and XII.
J.Med.Chem., 68, 2025
|
|
6GK7
 
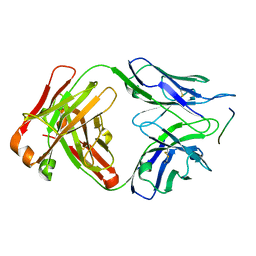 | | Crystal structure of anti-tau antibody dmCBTAU-27.1, double mutant (S31Y, T100I) of CBTAU-27.1, in complex with Tau peptide A8119B (residues 299-318) | | Descriptor: | CHLORIDE ION, HUMAN FAB ANTIBODY FRAGMENT OF CBTAU-27.1(S31Y,T100I), HUMAN TAU PEPTIDE A8119 RESIDUES 299-318 | | Authors: | Steinbacher, S, Mrosek, M, Juraszek, J. | | Deposit date: | 2018-05-18 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A common antigenic motif recognized by naturally occurring human VH5-51/VL4-1 anti-tau antibodies with distinct functionalities.
Acta Neuropathol Commun, 6, 2018
|
|
5GT4
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with (1R,2S,3R,5Z,7E,14beta,17alpha)-2-cyanopropoxy-9,10-secocholesta-5,7,10-triene-1,3,25-triol | | Descriptor: | 4-{[(1R,2S,3R,5Z,7E,14beta,17alpha)-1,3,25-trihydroxy-9,10-secocholesta-5,7,10-trien-2-yl]oxy}butanenitrile, Vitamin D3 receptor | | Authors: | Takimoto-Kamimura, M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the human vitamin D receptor ligand binding domain complexed with (1R,2S,3R,5Z,7E,14beta,17alpha)-2-cyanopropoxy-9,10-secocholesta-5,7,10-triene-1,3,25-triol
To Be Published
|
|
7AHX
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND D-ASPARTATE TENOFOVIR WITH BOUND MANGANESE | | Descriptor: | D-Aspartate Tenofovir, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(*TP*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-25 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
7AII
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND L-METHIONINE TENOFOVIR WITH BOUND MANGANESE | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), Gag-Pol polyprotein, ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
8TBS
 
 | | Structure of human erythrocyte pyruvate kinase in complex with an allosteric activator AG-946 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-[(6-aminopyridin-2-yl)methyl]-4-methyl-2-[(1H-pyrazol-3-yl)methyl]-4,6-dihydro-5H-[1,3]thiazolo[5',4':4,5]pyrrolo[2,3-d]pyridazin-5-one, MANGANESE (II) ION, ... | | Authors: | Jin, L, Padyana, A. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Based Design of AG-946, a Pyruvate Kinase Activator.
Chemmedchem, 19, 2024
|
|
