8CPI
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8CPJ
 
 | | Crystal structure of PPAR gamma (PPARG) in an inactive form | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
6F8U
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-20b | | Descriptor: | 2-[(~{E})-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]methylideneamino]oxy-1-[(2~{R},6~{R})-2,6-dimethylmorpholin-4-yl]ethanone, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6P80
 
 | | Structure of E. coli MS115-1 CdnC + ATP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Ye, Q, Azimi, C.S, Corbett, K.D. | | Deposit date: | 2019-06-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6P8V
 
 | | Structure of E. coli MS115-1 HORMA:CdnC:Trip13 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase, AAA family, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-08 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
8PMW
 
 | |
7A53
 
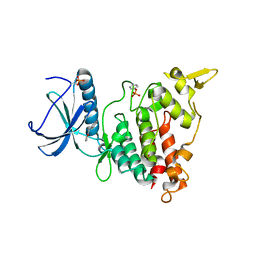 | | Structure of DYRK1A in complex with compound 7 | | Descriptor: | (3~{E})-3-[(5-methylfuran-2-yl)methylidene]-1~{H}-indol-2-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Hubbard, R.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Derived Selective Inhibitors of Dual-Specificity Kinases DYRK1A and DYRK1B.
J.Med.Chem., 64, 2021
|
|
8FV4
 
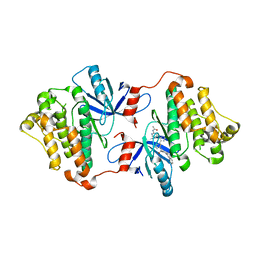 | | EGFR(T790M/V948R) in complex with compound 2 (LN5993) | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-{(3P)-3-[(4P)-4-(2-acetamidopyridin-4-yl)-2-(methylsulfanyl)-1H-imidazol-5-yl]phenyl}-11-oxo-10,11-dihydro-5H-dibenzo[b,e][1,4]diazepine-9-carboxamide, ... | | Authors: | Ogboo, B.C, Beyett, T.S, Eck, M.J, Heppner, D.E. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Linking ATP and allosteric sites to achieve superadditive binding with bivalent EGFR kinase inhibitors.
Commun Chem, 7, 2024
|
|
8FV3
 
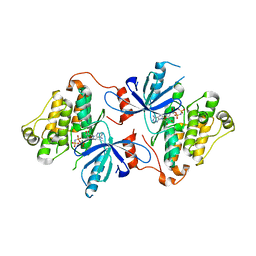 | | EGFR(T790M/V948R) in complex with compound 1 (LN4503) | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-{(4P)-4-[(4P)-5-{3-[(8-fluoro-11-oxo-5,11-dihydro-10H-dibenzo[b,e][1,4]diazepin-10-yl)methyl]phenyl}-2-(methylsulfanyl)-1H-imidazol-4-yl]pyridin-2-yl}acetamide, ... | | Authors: | Ogboo, B.C, Heppner, D.E. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Linking ATP and allosteric sites to achieve superadditive binding with bivalent EGFR kinase inhibitors.
Commun Chem, 7, 2024
|
|
6XL4
 
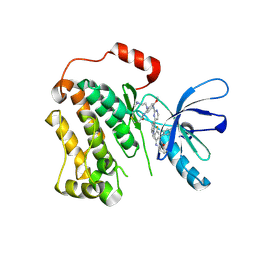 | | EGFR(T790M/V948R) in complex with AZD9291 and DDC4002 | | Descriptor: | 10-benzyl-8-fluoro-5,10-dihydro-11H-dibenzo[b,e][1,4]diazepin-11-one, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Heppner, D.E, Beyett, T.S, Eck, M.J. | | Deposit date: | 2020-06-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular basis for cooperative binding and synergy of ATP-site and allosteric EGFR inhibitors.
Nat Commun, 13, 2022
|
|
6GS4
 
 | | Crystal structure of peptide transporter DtpA-nanobody in complex with valganciclovir | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Dipeptide and tripeptide permease A, [(2~{S})-2-[(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)methoxy]-3-oxidanyl-propyl] (2~{S})-2-azanyl-3-methyl-butanoate, ... | | Authors: | Ural-Blimke, Y, Flayhan, A, Quistgaard, E.M, Loew, C. | | Deposit date: | 2018-06-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structure of Prototypic Peptide Transporter DtpA from E. coli in Complex with Valganciclovir Provides Insights into Drug Binding of Human PepT1.
J. Am. Chem. Soc., 141, 2019
|
|
8F3W
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) PAPAPAP variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | Schoenle, M.V, D'Andrea, E.D, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam antibiotics
To Be Published
|
|
8F3X
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) Poly-Gly variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | Schoenle, M.V, D'Andrea, E.D, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam antibiotics
To Be Published
|
|
8F3Y
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) Poly-Gly variant penicillin bound form from Enterococcus faecium | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin binding protein 5, SULFATE ION | | Authors: | Schoenle, M.V, D'Andrea, E.D, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam antibiotics
To Be Published
|
|
8RNE
 
 | | HLA-E*01:03 in complex with SARS-CoV-2 Nsp13 peptide, VMPLSAPTL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, E alpha chain variant, ... | | Authors: | Sun, R, Achour, A, Sala, B.M, Sandalova, T. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Emerging mutation in SARS-CoV-2 facilitates escape from NK cell recognition and associates with enhanced viral fitness.
Plos Pathog., 20, 2024
|
|
8F3V
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) PAPAPAP variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SULFATE ION | | Authors: | Schoenle, M.V, D'Andrea, E.D, Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam antibiotics
To Be Published
|
|
8RNF
 
 | | HLA-E*01:03 in complex with SARS-CoV-2 Omicron Nsp13 peptide, VIPLSAPTL | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, E alpha chain variant, ... | | Authors: | Sun, R, Achour, A, Sala, B.M, Sandalova, T. | | Deposit date: | 2024-01-09 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Emerging mutation in SARS-CoV-2 facilitates escape from NK cell recognition and associates with enhanced viral fitness.
Plos Pathog., 20, 2024
|
|
6X0C
 
 | | Tryptophan Synthase mutant beta-Q114A in complex with Cesium ion at the metal coordination site and aminoacrylate and benzimidazole at the enzyme beta site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BENZIMIDAZOLE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2020-05-15 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | External aldimine form of Tryptophan Synthase mutant beta-Q114A in complex with Cesium ion at the metal coordination site and benzimidazole at the enzyme beta site.
To be Published
|
|
8PMX
 
 | |
7SN7
 
 | |
8DO1
 
 | | Cryo-EM structure of the human Sec61 complex inhibited by ipomoeassin F | | Descriptor: | Protein transport protein Sec61 subunit alpha isoform 1, Protein transport protein Sec61 subunit beta, Protein transport protein Sec61 subunit gamma, ... | | Authors: | Park, E, Itskanov, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | A common mechanism of Sec61 translocon inhibition by small molecules.
Nat.Chem.Biol., 19, 2023
|
|
6G4M
 
 | | Torpedo californica acetylcholinesterase bound to uncharged hybrid reactivator 1 | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-[4-(1,2,3,4-tetrahydroacridin-9-ylamino)butyl]pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
6G4O
 
 | | Non-aged form of Torpedo californica acetylcholinesterase inhibited by tabun analog NEDPA bound to uncharged reactivator 1 | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-[4-(1,2,3,4-tetrahydroacridin-9-ylamino)butyl]pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
7AOK
 
 | | Crystal structure of CI2 mutant L49I | | Descriptor: | SULFATE ION, Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Olsen, J.G, Teilum, K, Hamborg, L, Roche, J.V. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Synergistic stabilization of a double mutant in chymotrypsin inhibitor 2 from a library screen in E. coli.
Commun Biol, 4, 2021
|
|
6ODC
 
 | | Crystal structure of HDAC8 in complex with compound 30 | | Descriptor: | (2E)-3-[2-(3-cyclopentyl-5,5-dimethyl-2-oxoimidazolidin-1-yl)phenyl]-N-hydroxyprop-2-enamide, 1,2-ETHANEDIOL, Histone deacetylase 8, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
