6ZK9
 
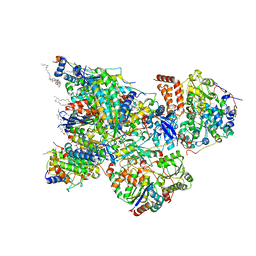 | | Peripheral domain of open complex I during turnover | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
7KG6
 
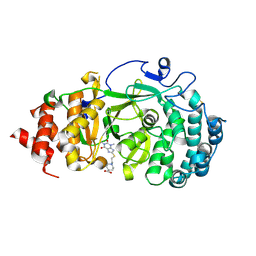 | | Structure of human PARG complexed with PARG-322 | | Descriptor: | 1-{2-[(1,3-dimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl)sulfanyl]ethyl}piperidine-4-carboxylic acid, Poly(ADP-ribose) glycohydrolase | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
6ZKC
 
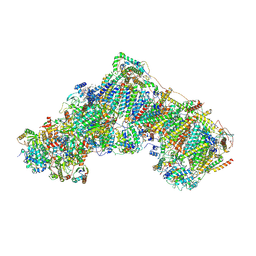 | | Complex I during turnover, closed | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6ZKR
 
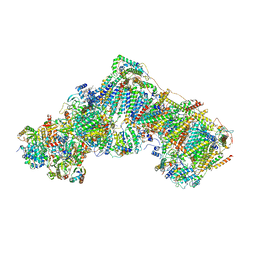 | | Native complex I, open3 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
7XKA
 
 | | Structure of human beta2 adrenergic receptor bound to constrained epinephrine | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Camelid Antibody Fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Xu, X, Shonberg, J, Kaindl, J, Clark, M, Stobel, A, Maul, L, Mayer, D, Hubner, H, Venkatakrishnan, A, Dror, R, Kobilka, B.K, Sunahara, R, Liu, X, Gmeiner, P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Constrained catecholamines gain beta 2 AR selectivity through allosteric effects on pocket dynamics.
Nat Commun, 14, 2023
|
|
7XK9
 
 | | Structure of human beta2 adrenergic receptor bound to constrained isoproterenol | | Descriptor: | (5R,6R)-6-(propan-2-ylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Camelid Antibody Fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Xu, X, Shonberg, J, Kaindl, J, Clark, M, Stobel, A, Maul, L, Mayer, D, Hubner, H, Venkatakrishnan, A, Dror, R, Kobilka, B.K, Sunahara, R, Liu, X, Gmeiner, P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Constrained catecholamines gain beta 2 AR selectivity through allosteric effects on pocket dynamics.
Nat Commun, 14, 2023
|
|
3HBQ
 
 | | Structure of recombinant Chicken Liver Sulfite Oxidase mutant Cys 185 Ala | | Descriptor: | GLYCEROL, HYDROXY(DIOXO)MOLYBDENUM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, ... | | Authors: | Qiu, J.A. | | Deposit date: | 2009-05-04 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of the C185S and C185A mutants of sulfite oxidase reveal rearrangement of the active site.
Biochemistry, 49, 2010
|
|
3HC2
 
 | | Crystal Structure of chicken sulfite oxidase mutant Tyr 322 Phe | | Descriptor: | MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, sulfite oxidase | | Authors: | Qiu, J.A. | | Deposit date: | 2009-05-05 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure function studies in sulfite oxidase with altered active sites
To be Published
|
|
3WQJ
 
 | | Crystal structure of archaerhodopsin-2 at 1.8 angstrom resolution | | Descriptor: | (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, ... | | Authors: | Kouyama, T. | | Deposit date: | 2014-01-27 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of archaerhodopsin-2 at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LUW
 
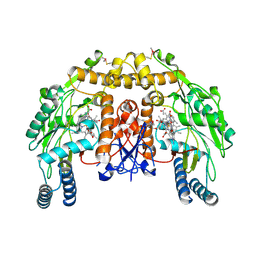 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 6-((((3R,5S)-5-(((6-amino-4-methylpyridin-2-yl)methoxy)methyl)pyrrolidin-3-yl)oxy)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-({[(3R,5S)-5-{[(6-amino-4-methylpyridin-2-yl)methoxy]methyl}pyrrolidin-3-yl]oxy}methyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-07-25 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and biological studies on bacterial nitric oxide synthase inhibitors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5UEN
 
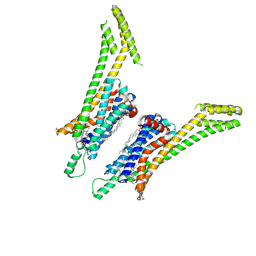 | | Crystal structure of the human adenosine A1 receptor A1AR-bRIL in complex with the covalent antagonist DU172 at 3.2A resolution | | Descriptor: | 4-{[3-(8-cyclohexyl-2,6-dioxo-1-propyl-1,2,6,7-tetrahydro-3H-purin-3-yl)propyl]carbamoyl}benzene-1-sulfonyl fluoride, Adenosine receptor A1,Soluble cytochrome b562,Adenosine receptor A1, OLEIC ACID | | Authors: | Glukhova, A, Thal, D.M, Nguyen, A.T, Vecchio, E.A, Jorg, M, Scammells, P.J, May, L.T, Sexton, P.M, Christopoulos, A. | | Deposit date: | 2017-01-03 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Adenosine A1 Receptor Reveals the Basis for Subtype Selectivity.
Cell, 168, 2017
|
|
7KB8
 
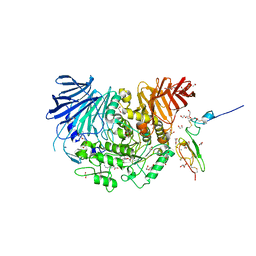 | | Co-crystal structure of alpha glucosidase with compound 8 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(4-{[2-nitro-4-(triazan-1-yl)phenyl]amino}butyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-10-01 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7XWW
 
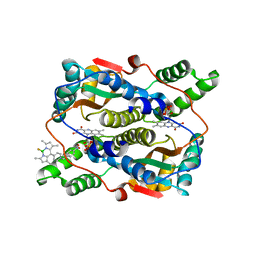 | | Crystal structure of NTR in complex with BN-XB | | Descriptor: | 2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-8-[1-[(4-nitrophenyl)methyl]pyridin-1-ium-4-yl]-3-aza-1-azonia-2-boranuidatricyclo[7.3.0.0^{3,7}]dodeca-1(12),4,6,8,10-pentaene, Dihydropteridine reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Chen, X, Chen, J, Li, J.L. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of NTR in complex with BN-XB
To Be Published
|
|
4LXA
 
 | | Crystal Structure of Human Beta Secretase in Complex with Compound 11a | | Descriptor: | (1R,3S,4S,5R)-3-{4-amino-3-fluoro-5-[(1,1,1,3,3,3-hexafluoropropan-2-yl)oxy]benzyl}-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1-oxide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2013-07-29 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of cyclic sulfoxide hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3HHK
 
 | | HCV NS5b polymerase complex with a substituted benzothiadizine | | Descriptor: | 2-({(3R)-3-[(3S)-1-(3-methylbutyl)-2,4-dioxo-1,2,3,4-tetrahydroquinolin-3-yl]-1,1-dioxido-3,4-dihydro-2H-1,2,4-benzothiadiazin-7-yl}oxy)acetamide, HCV NS5 polymerase | | Authors: | Concha, N.O, Singh, O. | | Deposit date: | 2009-05-15 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substituted benzothiadizine inhibitors of Hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3KI4
 
 | | Catalytic fragment of Cholix toxin from Vibrio Cholerae in complex with inhibitor GP-P | | Descriptor: | (11bR)-3-oxo-1,2,3,11b-tetrahydrochromeno[4,3,2-de]isoquinoline-10-sulfonic acid, Cholix toxin, GLYCEROL | | Authors: | Jorgensen, R, Edwards, P.R, Merrill, A.R. | | Deposit date: | 2009-10-31 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure function analysis of soluble inhibitors of cholix toxin from Vibrio cholerae
To be Published
|
|
4LZR
 
 | | Crystal Structure of BRD4(1) bound to Colchicine | | Descriptor: | Bromodomain-containing protein 4, N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wohlwend, D, Gerhardt, S, Einsle, O, Huegle, M. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
5Q1H
 
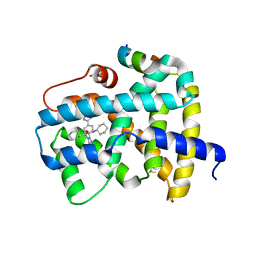 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-{2-[4-(1H-tetrazol-5-yl)phenyl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
7KVZ
 
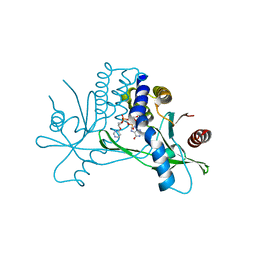 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-2 | | Descriptor: | (2R,5R,7R,8R,10S,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2,10,16-trihydroxy-14-[(pyrimidin-4-yl)oxy]decahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R.J. | | Deposit date: | 2020-11-29 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
3WZU
 
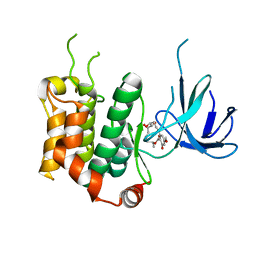 | | THE STRUCTURE OF MAP2K7 IN COMPLEX WITH 5Z-7-oxozeaenol | | Descriptor: | (3S,5Z,8S,9S,11E)-8,9,16-trihydroxy-14-methoxy-3-methyl-3,4,9,10-tetrahydro-1H-2-benzoxacyclotetradecine-1,7(8H)-dione, Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Sogabe, Y, Hashimoto, Y, Matsumoto, T, Kinoshita, T. | | Deposit date: | 2014-10-07 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | 5Z-7-Oxozeaenol covalently binds to MAP2K7 at Cys218 in an unprecedented manner.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7KPF
 
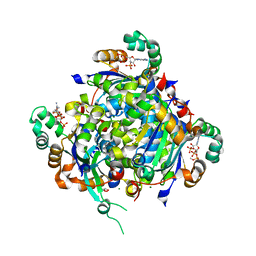 | | NME2 bound to myristoyl-CoA | | Descriptor: | GLYCEROL, MAGNESIUM ION, Nucleoside diphosphate kinase B, ... | | Authors: | Price, I.R, Lin, H. | | Deposit date: | 2020-11-11 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A Chemical Proteomic Approach reveals the regulation of NME1/2 and cancer metastasis by Long-Chain Fatty Acyl Coenzyme A
To Be Published
|
|
9F0R
 
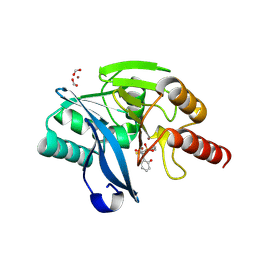 | | VIM-2 in complex with GKV65 (5g) - dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors | | Descriptor: | FORMIC ACID, MAGNESIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Bosman, R, Prester, A, Bartels, K, Gulyas, K.V, Erdelyi, M, Schulz, E.C. | | Deposit date: | 2024-04-17 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Dynamically chiral phosphonic acid-type metallo-beta-lactamase inhibitors.
Commun Chem, 8, 2025
|
|
3KI6
 
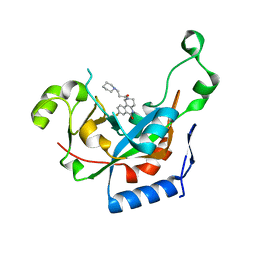 | | Catalytic fragment of Cholix toxin from Vibrio Cholerae in complex with inhibitor GP-L | | Descriptor: | 8-fluoro-2-(3-piperidin-1-ylpropanoyl)-1,3,4,5-tetrahydrobenzo[c][1,6]naphthyridin-6(2H)-one, Cholix toxin | | Authors: | Jorgensen, R, Edwards, P.R, Merrill, A.R. | | Deposit date: | 2009-10-31 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure function analysis of soluble inhibitors of cholix toxin from Vibrio cholerae
To be Published
|
|
5UIM
 
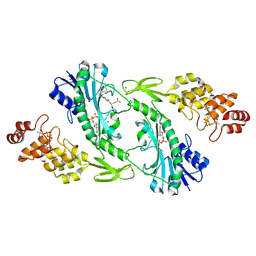 | | X-ray structure of the FdtF N-formyltransferase from salmonella enteric O60 in complex with folinic acid and TDP-Qui3N | | Descriptor: | Formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, POTASSIUM ION, ... | | Authors: | Woodford, C.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
6CF7
 
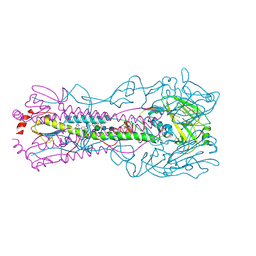 | |
