7SI4
 
 | | CRYSTAL STRUCTURE OF EED WITH MRTX-2219 | | Descriptor: | (4S)-8-{4-[(dimethylamino)methyl]-2-methylphenyl}-5-{[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]amino}imidazo[1,2-c]pyrimidine-2-carbonitrile, FORMIC ACID, Polycomb protein EED | | Authors: | Gunn, R.J, Burns, A.C, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF EED WITH MRTX-2219
TO BE PUBLISHED
|
|
1BX6
 
 | | CRYSTAL STRUCTURE OF THE POTENT NATURAL PRODUCT INHIBITOR BALANOL IN COMPLEX WITH THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE | | Descriptor: | BALANOL, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Xuong, N.-H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1998-10-13 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the potent natural product inhibitor balanol in complex with the catalytic subunit of cAMP-dependent protein kinase.
Biochemistry, 38, 1999
|
|
1BKX
 
 | | A BINARY COMPLEX OF THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE AND ADENOSINE FURTHER DEFINES CONFORMATIONAL FLEXIBILITY | | Descriptor: | ADENOSINE MONOPHOSPHATE, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Cox, S, Xuong, N, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1997-07-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A binary complex of the catalytic subunit of cAMP-dependent protein kinase and adenosine further defines conformational flexibility.
Structure, 5, 1997
|
|
2OZA
 
 | | Structure of p38alpha complex | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14 | | Authors: | White, A, Pargellis, C.A, Studts, J.M, Werneburg, B.G, Farmer II, B.T. | | Deposit date: | 2007-02-25 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of MAPK-activated protein kinase 2:p38 assembly
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1D2N
 
 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | Authors: | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1998-06-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
4BTE
 
 | | DJ-1 Cu(I) complex | | Descriptor: | COPPER (I) ION, PROTEIN DJ-1 | | Authors: | Puno, M.R.A, Odell, M, Moody, P.C.E. | | Deposit date: | 2013-06-14 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of Cu(I)-Bound Dj-1 Reveals a Biscysteinate Metal Binding Site at the Homodimer Interface: Insights Into Mutational Inactivation of Dj-1 in Parkinsonism.
J.Am.Chem.Soc., 135, 2013
|
|
4HR9
 
 | | Human interleukin 17A | | Descriptor: | Interleukin-17A | | Authors: | Liu, S. | | Deposit date: | 2012-10-26 | | Release date: | 2013-05-22 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structures of interleukin 17A and its complex with IL-17 receptor A.
Nat Commun, 4, 2013
|
|
2PUU
 
 | |
1ERT
 
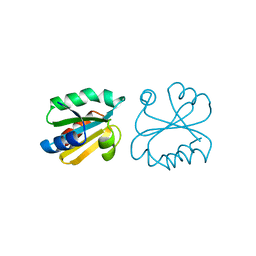 | | HUMAN THIOREDOXIN (REDUCED FORM) | | Descriptor: | THIOREDOXIN | | Authors: | Weichsel, A, Gasdaska, J.R, Powis, G, Montfort, W.R. | | Deposit date: | 1996-02-07 | | Release date: | 1996-10-14 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of reduced, oxidized, and mutated human thioredoxins: evidence for a regulatory homodimer.
Structure, 4, 1996
|
|
1UBD
 
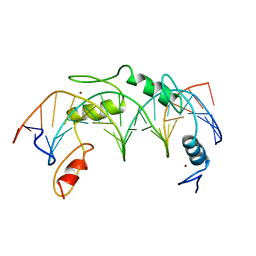 | | CO-CRYSTAL STRUCTURE OF HUMAN YY1 ZINC FINGER DOMAIN BOUND TO THE ADENO-ASSOCIATED VIRUS P5 INITIATOR ELEMENT | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*CP*TP*CP*CP*AP*TP*TP*TP*TP*GP*AP*A P*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*CP*AP*AP*AP*AP*TP*GP*GP*AP*GP*AP*C P*CP*CP*T)-3'), PROTEIN (YY1 ZINC FINGER DOMAIN), ... | | Authors: | Houbaviy, H.B, Usheva, A, Shenk, T, Burley, S.K. | | Deposit date: | 1996-10-04 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
2E70
 
 | | Solution structure of the fifth KOW motif of human transcription elongation factor SPT5 | | Descriptor: | Transcription elongation factor SPT5 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-05 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fifth KOW motif of human transcription elongation factor SPT5
To be Published
|
|
6IN8
 
 | | Crystal structure of MucB | | Descriptor: | Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6V0N
 
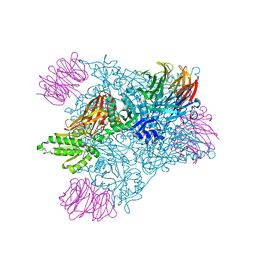 | | PRMT5 bound to PBM peptide from Riok1 | | Descriptor: | Methylosome protein 50, Protein arginine N-methyltransferase 5, Riok1 PBM peptide, ... | | Authors: | McMIllan, B.J, Raymond, D.D. | | Deposit date: | 2019-11-19 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mol.Cell, 81, 2021
|
|
6UXY
 
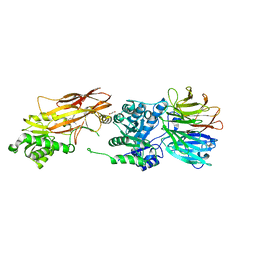 | | PRMT5:MEP50 Complexed with Allosteric Inhibitor Compound 8 | | Descriptor: | (5R)-2-amino-5-(2-cyclohexylethyl)-3-methyl-5-phenyl-3,5-dihydro-4H-imidazol-4-one, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Palte, R.L, Schneider, S.E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5).
Acs Med.Chem.Lett., 11, 2020
|
|
8G0H
 
 | |
6IN9
 
 | | Crystal structure of MucB in complex with MucA(peri) | | Descriptor: | Sigma factor AlgU negative regulatory protein, Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6V0O
 
 | | PRMT5 bound to the PBM peptide from pICln | | Descriptor: | ACETYL GROUP, Methylosome protein 50, PBM peptide, ... | | Authors: | McMillan, B.J, Raymond, D.D. | | Deposit date: | 2019-11-19 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mol.Cell, 81, 2021
|
|
3L5X
 
 | | Crystal structure of the complex between IL-13 and H2L6 FAB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, H2L6 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
6UXX
 
 | | PRMT5:MEP50 Complexed with Allosteric Inhibitor Compound 1a | | Descriptor: | (5R)-2-amino-5-(4-methoxyphenyl)-3-methyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-yl]-3,5-dihydro-4H-imidazol-4-one, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Palte, R.L, Schneider, S.E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5).
Acs Med.Chem.Lett., 11, 2020
|
|
3LL8
 
 | |
6V0P
 
 | | PRMT5 complex bound to covalent PBM inhibitor BRD6711 | | Descriptor: | 2-(5-chloro-6-oxopyridazin-1(6H)-yl)-N-(4-methyl-3-sulfamoylphenyl)acetamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | McMillan, B.J, McKinney, D.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the PRMT5-Substrate Adaptor Interaction.
J.Med.Chem., 64, 2021
|
|
3L5W
 
 | | Crystal structure of the complex between IL-13 and C836 FAB | | Descriptor: | C836 HEAVY CHAIN, C836 LIGHT CHAIN, GLYCEROL, ... | | Authors: | Teplyakov, A, Obmolova, G, Malia, T, Gilliland, G.L. | | Deposit date: | 2009-12-22 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human framework adaptation of a mouse anti-human IL-13 antibody.
J.Mol.Biol., 398, 2010
|
|
7VG7
 
 | | Plexin B1 extracellular fragment in complex with lasso-grafted PB1m6A9 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-B1, TRIETHYLENE GLYCOL, ... | | Authors: | Sugano, N.N, Hirata, K, Yamashita, K, Yamamoto, M, Arimori, T, Takagi, J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo Fc-based receptor dimerizers differentially modulate PlexinB1 function.
Structure, 30, 2022
|
|
7VF3
 
 | | Plexin B1 extracellular fragment in complex with lasso-grafted PB1m7 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Plexin-B1, ... | | Authors: | Sugano, N.N, Hirata, K, Yamashita, K, Yamamoto, M, Arimori, T, Takagi, J. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | De novo Fc-based receptor dimerizers differentially modulate PlexinB1 function.
Structure, 30, 2022
|
|
3QFA
 
 | | Crystal structure of the human thioredoxin reductase-thioredoxin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Thioredoxin, ... | | Authors: | Fritz-Wolf, K, Kehr, S, Stumpf, M, Rahlfs, S, Becker, K. | | Deposit date: | 2011-01-21 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human thioredoxin reductase-thioredoxin complex
Nat Commun, 2, 2011
|
|
