5O77
 
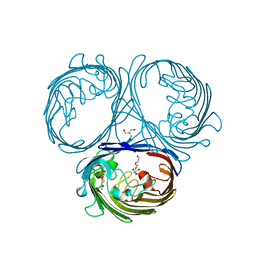 | | Klebsiella pneumoniae OmpK35 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, OmpK35 | | Authors: | van den berg, B, Pathania, M, Zahn, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Getting Drugs into Gram-Negative Bacteria: Rational Rules for Permeation through General Porins.
Acs Infect Dis., 4, 2018
|
|
5O7M
 
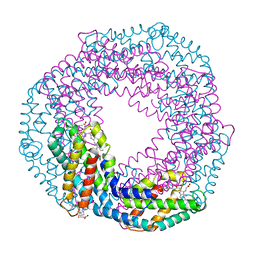 | | Single-shot pink beam serial crystallography: Phycocyanin (One chip, chip_1) | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V, Sarrou, I. | | Deposit date: | 2017-06-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
5UZB
 
 | | Cryo-EM structure of the MAL TIR domain filament | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Ve, T, Vajjhala, P.R, Hedger, A, Croll, T, DiMaio, F, Horsefield, S, Yu, X, Lavrencic, P, Hassan, Z, Morgan, G.P, Mansell, A, Mobli, M, O'Carrol, A, Chauvin, B, Gambin, Y, Sierecki, E, Landsberg, M.J, Stacey, K.J, Egelman, E.H, Kobe, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-07-26 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of TIR-domain-assembly formation in MAL- and MyD88-dependent TLR4 signaling.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5UYZ
 
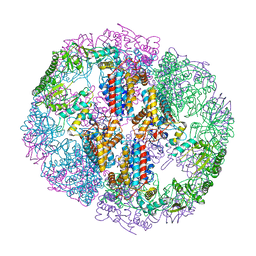 | | Structure of Human T-complex protein 1 subunit epsilon (CCT5) mutant His147Arg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T-complex protein 1 subunit epsilon | | Authors: | Pereira, J.H, McAndrew, R.P, Sergeeva, O.A, Ralston, C.Y, King, J.A, Adams, P.D. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the human TRiC/CCT Subunit 5 associated with hereditary sensory neuropathy.
Sci Rep, 7, 2017
|
|
5V1M
 
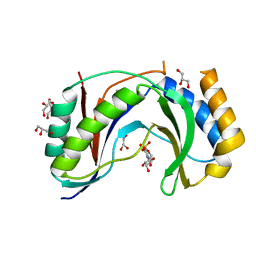 | | Structure of human Usb1 with uridine 5'-monophosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, U6 snRNA phosphodiesterase, ... | | Authors: | Montemayor, E.J, Didychuk, A.L, Butcher, S.E. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Usb1 controls U6 snRNP assembly through evolutionarily divergent cyclic phosphodiesterase activities.
Nat Commun, 8, 2017
|
|
5URW
 
 | |
5UYX
 
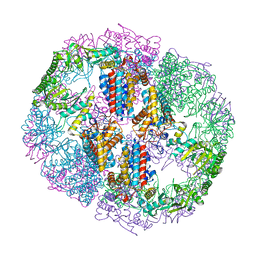 | | Structure of Human T-complex protein 1 subunit epsilon (CCT5) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit epsilon | | Authors: | Pereira, J.H, McAndrew, R.P, Sergeeva, O.A, Ralston, C.Y, King, J.A, Adams, P.D. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the human TRiC/CCT Subunit 5 associated with hereditary sensory neuropathy.
Sci Rep, 7, 2017
|
|
3TOK
 
 | | Assaying the energies of biological halogen bonds. | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*TP*AP*CP*CP*GP*G)-3'), DNA (5'-D(*CP*CP*GP*GP*TP*AP*TP*CP*GP*G)-3'), SODIUM ION | | Authors: | Carter, M, Ho, P.S. | | Deposit date: | 2011-09-05 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Assaying the Energies of Biological Halogen Bonds
CRYST.GROWTH DES., 11, 2011
|
|
5VCM
 
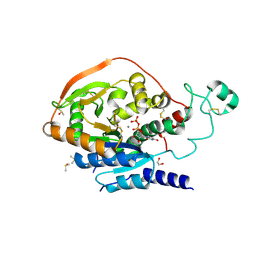 | | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase with bound UDP and Manganese | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase, ... | | Authors: | Sanders, J.H, Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | HumanN-acetylglucosaminyltransferase II substrate recognition uses a modular architecture that includes a convergent exosite.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5UTK
 
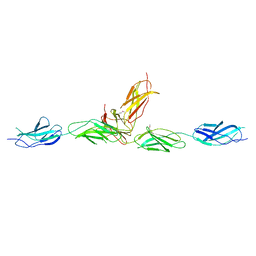 | |
5UAS
 
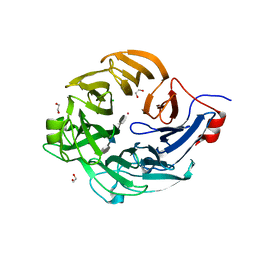 | | Structure of a new family of Polysaccharide lyase PL25-Ulvanlyase bound to -[GlcA(1-4)Rha3S]- | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CHLORIDE ION, ... | | Authors: | Ulaganathan, T.S, Cygler, M. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Ulvan-Degrading Polysaccharide Lyase Family: Structure and Catalytic Mechanism Suggests Convergent Evolution of Active Site Architecture.
ACS Chem. Biol., 12, 2017
|
|
5TIE
 
 | | x-ray structure of acyl-CoA thioesterase I, TesA, mutant M141L/Y145K/L146K at pH 7.5 in complex with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
3SLM
 
 | | Crystal structure of the 2'- Deoxyguanosine riboswitch bound to 2'-deoxyguanosine-5'-monophosphate | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, RNA (68-MER), ... | | Authors: | Pikovskaya, O, Polonskaia, A, Patel, D.J, Serganov, A.A. | | Deposit date: | 2011-06-24 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural principles of nucleoside selectivity in a 2'-deoxyguanosine riboswitch.
Nat.Chem.Biol., 7, 2011
|
|
5T2Z
 
 | | Crystal Structure of Multi-drug Resistant HIV-1 Protease PR-S17 in Complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-08-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Studies of a Rationally Selected Multi-Drug Resistant HIV-1 Protease Reveal Synergistic Effect of Distal Mutations on Flap Dynamics.
PLoS ONE, 11, 2016
|
|
5T4Y
 
 | | Crystal structure of BT1762-1763 | | Descriptor: | MAGNESIUM ION, SusC homolog, SusD homolog | | Authors: | van den Berg, B. | | Deposit date: | 2016-08-30 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
3SUP
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dCTP Opposite 2AP (GC rich sequence) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
5VCR
 
 | | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase with bound uranium dioxide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase, CHLORIDE ION, ... | | Authors: | Sanders, J.H, Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | HumanN-acetylglucosaminyltransferase II substrate recognition uses a modular architecture that includes a convergent exosite.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OPY
 
 | | Crystal structure of anti-alphaVbeta3 integrin Fab LM609 | | Descriptor: | Heavy chain of LM609 Fab (antigen-binding fragment), Light chain of LM609 Fab (antigen-binding fragment) | | Authors: | Backovic, M, Veesler, D, Borst, A.J, James, Z.M, Zagotta, W, Ginsberg, M, Rey, F.A, DiMaio, F. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human alpha V beta 3 Integrin via Steric Hindrance.
Structure, 25, 2017
|
|
5VI4
 
 | | IL-33/ST2/IL-1RAcP ternary complex structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein, Interleukin-1 receptor-like 1, ... | | Authors: | Guenther, S, Sundberg, E.J. | | Deposit date: | 2017-04-13 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | IL-1 Family Cytokines Use Distinct Molecular Mechanisms to Signal through Their Shared Co-receptor.
Immunity, 47, 2017
|
|
5VRK
 
 | | Crystal structure of SsoPox AsA6 mutant (F46L-C258A-W263M-I280T) - open form | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Hiblot, J, Gotthard, G, Jacquet, P, Daude, D, Bergonzi, C, Chabriere, E, Elias, M. | | Deposit date: | 2017-05-10 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rational engineering of a native hyperthermostable lactonase into a broad spectrum phosphotriesterase.
Sci Rep, 7, 2017
|
|
5W1V
 
 | | Structure of the HLA-E-VMAPRTLIL/GF4 TCR complex | | Descriptor: | Beta-2-microglobulin, GF4 T cell receptor alpha chain, GF4 T cell receptor beta chain, ... | | Authors: | Gras, S, Walpole, N, Farenc, C, Rossjohn, J. | | Deposit date: | 2017-06-04 | | Release date: | 2017-10-04 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | A conserved energetic footprint underpins recognition of human leukocyte antigen-E by two distinct alpha beta T cell receptors.
J. Biol. Chem., 292, 2017
|
|
5VM5
 
 | | Engineered tryptophan synthase b-subunit from Pyrococcus furiosus, PfTrpB2B9, with Ser bound | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, SODIUM ION, Tryptophan synthase beta chain 1, ... | | Authors: | Buller, A.R, van Roye, P. | | Deposit date: | 2017-04-26 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Directed Evolution Mimics Allosteric Activation by Stepwise Tuning of the Conformational Ensemble.
J. Am. Chem. Soc., 140, 2018
|
|
3SUN
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dTTP Opposite 2AP (AT rich sequence) | | Descriptor: | 5'-D(*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*AP*AP*TP*TP*AP*AP*TP*TP*AP*AP*TP*TP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
5V7I
 
 | | Crystal structure of homo sapiens serine hydroxymethyltransferase 2 (mitochondrial) (SHMT2), in complex with glycine, PLP and folate-competitive pyrazolopyran inhibitor: 6-amino-4-isopropyl-3-methyl-4-(3-(pyrrolidin-1-yl)-5-(trifluoromethyl)phenyl)-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile | | Descriptor: | (4R)-6-amino-3-methyl-4-(propan-2-yl)-4-[3-(pyrrolidin-1-yl)-5-(trifluoromethyl)phenyl]-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ducker, G.S, Ghergurovich, J.M, Mainolfi, N, Suri, V, Jeong, S, Friedman, A, Manfredi, M, Kim, H, Rabinowitz, J.D. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3SUO
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dTTP Opposite 2AP (GC rich sequence) | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*GP*GP*CP*GP*GP*CP*GP*(2DA))-3', 5'-D(P*CP*(2PR)P*TP*CP*GP*CP*CP*GP*CP*CP*GP*CP*GP*CP*GP*G)-3', CALCIUM ION, ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-07-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of the 2-Aminopurine-Cytosine Base Pair Formed in the Polymerase Active Site of the RB69 Y567A-DNA Polymerase.
Biochemistry, 50, 2011
|
|
