7MXO
 
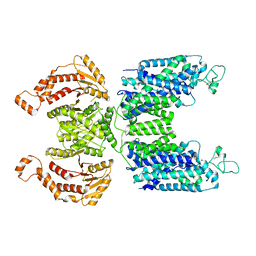 | | CryoEM structure of human NKCC1 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Solute carrier family 12 member 2 | | Authors: | Moseng, M.A. | | Deposit date: | 2021-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
6EQE
 
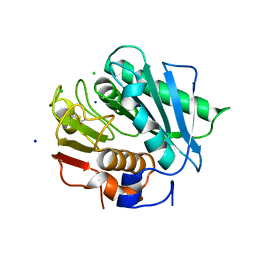 | | High resolution crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SODIUM ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5B5F
 
 | | Crystal structure of ALiS3-Streptavidin complex | | Descriptor: | N-methyl-3-(4-oxo-4,5-dihydrofuro[3,2-c]pyridin-2-yl)benzenesulfonamide, Streptavidin | | Authors: | Sugiyama, S, Terai, T, Kakinouchi, K, Fujikake, R, Nagano, T, Urano, Y. | | Deposit date: | 2016-05-04 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Improving the Solubility of Artificial Ligands of Streptavidin to Enable More Practical Reversible Switching of Protein Localization in Cells
Chembiochem, 18, 2017
|
|
5AW9
 
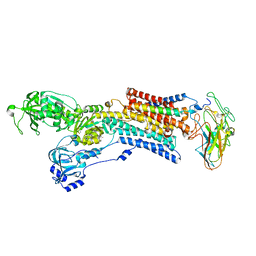 | | Kinetics by X-ray crystallography: native E2.MgF42-.2K+ crystal for Rb+ bound crystals | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
6EL8
 
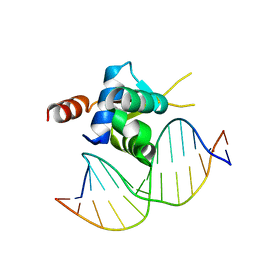 | | Crystal structure of the Forkhead domain of human FOXN1 in complex with DNA | | Descriptor: | DNA (5'-D(*GP*GP*TP*GP*GP*CP*GP*TP*CP*TP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*AP*GP*AP*CP*GP*CP*CP*AP*CP*C)-3'), Forkhead box protein N1 | | Authors: | Newman, J.A, Aitkenhead, H.A, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of the Forkhead domain of human FOXN1 in complex with DNA
To be published
|
|
6ELI
 
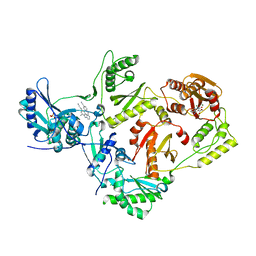 | | Structure of HIV-1 reverse transcriptase (RT) in complex with rilpivirine and an RNase H inhibitor XZ462 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Gag-Pol polyprotein, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2017-09-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Developing and Evaluating Inhibitors against the RNase H Active Site of HIV-1 Reverse Transcriptase.
J. Virol., 92, 2018
|
|
6EB6
 
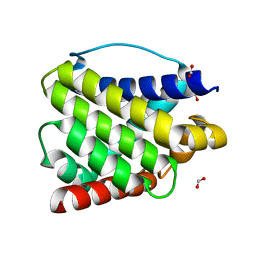 | | Crystal structure of BAX W139A monomer | | Descriptor: | Apoptosis regulator BAX, FORMIC ACID | | Authors: | Robin, A.Y. | | Deposit date: | 2018-08-05 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | BAX Activation: Mutations Near Its Proposed Non-canonical BH3 Binding Site Reveal Allosteric Changes Controlling Mitochondrial Association.
Cell Rep, 27, 2019
|
|
6EBK
 
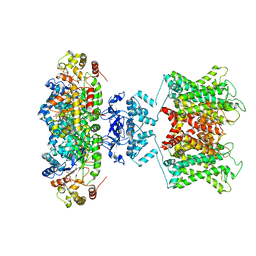 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6EOG
 
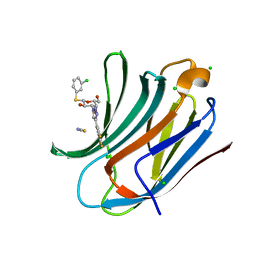 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{R},6~{R})-2-(3-chlorophenyl)sulfanyl-6-(hydroxymethyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, CHLORIDE ION, Galectin-3, ... | | Authors: | Hakansson, M, Nilsson, U.J, Zetterberg, F, Logan, D.T. | | Deposit date: | 2017-10-09 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Monosaccharide Derivatives with Low-Nanomolar Lectin Affinity and High Selectivity Based on Combined Fluorine-Amide, Phenyl-Arginine, Sulfur-pi , and Halogen Bond Interactions.
ChemMedChem, 13, 2018
|
|
5B1B
 
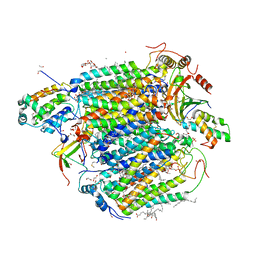 | | Bovine heart cytochrome c oxidase in the fully reduced state at 1.6 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Yano, N, Muramoto, K, Shimada, A, Takemura, S, Baba, J, Fujisawa, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2015-12-01 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Mg2+-containing Water Cluster of Mammalian Cytochrome c Oxidase Collects Four Pumping Proton Equivalents in Each Catalytic Cycle.
J.Biol.Chem., 291, 2016
|
|
5B3H
 
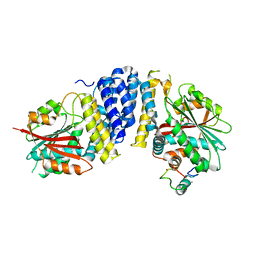 | | The crystal structure of the JACKDAW/IDD10 bound to the heterodimeric SHR-SCR complex | | Descriptor: | Protein SCARECROW, Protein SHORT-ROOT, ZINC ION, ... | | Authors: | Hirano, Y, Suyama, T, Nakagawa, M, Hakoshima, T. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD
Nat Plants, 3, 2017
|
|
7MIT
 
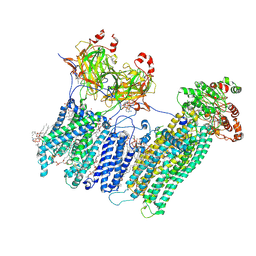 | | Vascular KATP channel: Kir6.1 SUR2B propeller-like conformation 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ... | | Authors: | Sung, M.W, Shyng, S.L. | | Deposit date: | 2021-04-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Vascular K ATP channel structural dynamics reveal regulatory mechanism by Mg-nucleotides.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5AVU
 
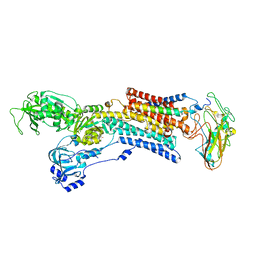 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 7.0 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
5AP6
 
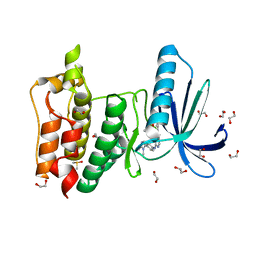 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DUAL SPECIFICITY PROTEIN KINASE TTK, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.M, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
5AW2
 
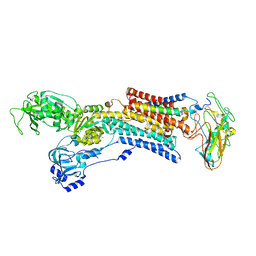 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 85 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
6ECL
 
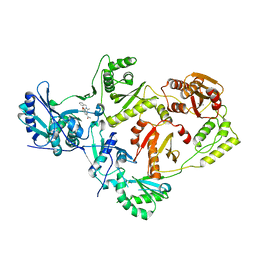 | | Crystal Structure of a 1,2,4-Triazole Allosteric RNase H Inhibitor in Complex with HIV Reverse Transcriptase | | Descriptor: | 1-[4-methoxy-3-[[5-methyl-4-(phenylmethyl)-1,2,4-triazol-3-yl]sulfanylmethyl]phenyl]ethanone, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Crystal Structure of a 1,2,4-Triazole Allosteric RNase H Inhibitor in Complex with HIV Reverse Transcriptase
To be published
|
|
6EMI
 
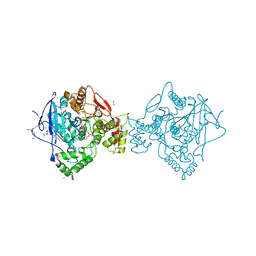 | | Crystal structure of a variant of human butyrylcholinesterase expressed in bacteria. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cholinesterase, ... | | Authors: | Brazzolotto, X, Igert, A, Guillon, V, Santoni, G, Nachon, F. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.476 Å) | | Cite: | Bacterial Expression of Human Butyrylcholinesterase as a Tool for Nerve Agent Bioscavengers Development.
Molecules, 22, 2017
|
|
5B3Z
 
 | |
6EHJ
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and peptide bound | | Descriptor: | ASPARAGINE, COENZYME A, GLYCEROL, ... | | Authors: | Perez-Dorado, I, Ritzefeld, M, Tate, E.W. | | Deposit date: | 2017-09-13 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation.
Nat Commun, 11, 2020
|
|
5B6X
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 760 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
6EQF
 
 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup P212121 | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5BKC
 
 | | Crystal structure of AAD-1 in complex with (R)-diclofop, Mn(II), and 2-oxoglutarate | | Descriptor: | (2R)-2-{4-[(3,5-dichloropyridin-2-yl)oxy]phenoxy}propanoic acid, (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, 2-OXOGLUTARIC ACID, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2019-06-02 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5BK9
 
 | | AAD-1 Bound to the Vanadyl Ion and Succinate | | Descriptor: | (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, PHOSPHATE ION, SUCCINIC ACID, ... | | Authors: | Ongpipattanakul, C, Chekan, J.R. | | Deposit date: | 2019-06-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular basis for enantioselective herbicide degradation imparted by aryloxyalkanoate dioxygenases in transgenic plants.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7NEP
 
 | | Homology model of the in situ actomyosin complex from the A-band of mouse psoas muscle sarcomere in the rigor state | | Descriptor: | Actin, alpha skeletal muscle, Myosin light chain 1/3, ... | | Authors: | Wang, Z, Grange, M, Wagner, T, Kho, A.L, Gautel, M, Raunser, S. | | Deposit date: | 2021-02-04 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (10.2 Å) | | Cite: | The molecular basis for sarcomere organization in vertebrate skeletal muscle.
Cell, 184, 2021
|
|
5AVW
 
 | | Kinetics by X-ray crystallography: Tl+-substitution of bound K+ in the E2.MgF42-.2K+ crystal after 16.5 min | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Ogawa, H, Cornelius, F, Hirata, A, Toyoshima, C. | | Deposit date: | 2015-07-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequential substitution of K(+) bound to Na(+),K(+)-ATPase visualized by X-ray crystallography.
Nat Commun, 6, 2015
|
|
