4ZAD
 
 | | Structure of C. dubliensis Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Fdc1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
7C6J
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellobiose | | Descriptor: | ACETATE ION, CHLORIDE ION, SULFUR DIOXIDE, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
4ZAV
 
 | | UbiX in complex with a covalent adduct between dimethylallyl monophosphate and reduced FMN | | Descriptor: | 1-deoxy-1-[7,8-dimethyl-5-(3-methylbut-2-en-1-yl)-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono -D-ribitol, PHOSPHATE ION, SODIUM ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
4G8B
 
 | |
4ZA9
 
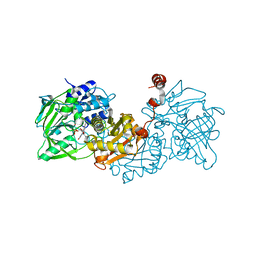 | | Structure of A. niger fdc1 in complex with a phenylpyruvate derived adduct to the prenylated flavin cofactor | | Descriptor: | 1-deoxy-5-O-phosphono-1-[(1S)-3,3,4,5-tetramethyl-9,11-dioxo-1-(phenylacetyl)-2,3,8,9,10,11-hexahydro-1H,7H-quinolino[1 ,8-fg]pteridin-7-yl]-D-ribitol, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
7C7V
 
 | | Vitamin D3 receptor/lithochoric acid derivative complex | | Descriptor: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, FORMIC ACID, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Masuno, H, Numoto, N, Kagechika, H, Ito, N. | | Deposit date: | 2020-05-26 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lithocholic Acid Derivatives as Potent Vitamin D Receptor Agonists.
J.Med.Chem., 64, 2021
|
|
5ZTC
 
 | |
4G90
 
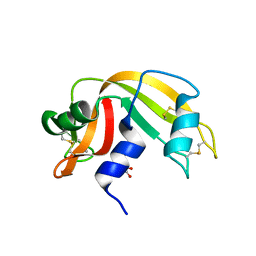 | | Crystal structure of Ribonuclease A in complex with 5e | | Descriptor: | 1-{[1-(alpha-L-arabinofuranosyl)-1H-1,2,3-triazol-4-yl]methyl}-5-fluoro-2,4-dioxo-1,2,3,4-tetrahydropyrimidine, Ribonuclease pancreatic | | Authors: | Chatzileontiadou, D.S.M, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Triazole pyrimidine nucleosides as inhibitors of Ribonuclease A. Synthesis, biochemical, and structural evaluation.
Bioorg.Med.Chem., 20, 2012
|
|
4Z34
 
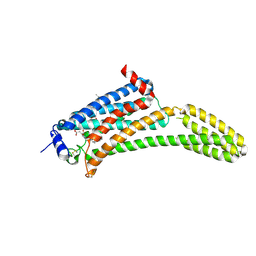 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO9780307 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, Lysophosphatidic acid receptor 1, Soluble cytochrome b562, ... | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4Z3Y
 
 | | Active site complex BamBC of Benzoyl Coenzyme A reductase in complex with Benzoyl-CoA | | Descriptor: | Benzoyl-CoA reductase, putative, IRON/SULFUR CLUSTER, ... | | Authors: | Weinert, T, Kung, J.W, Weidenweber, S, Huwiler, S.G, Boll, M, Ermler, U. | | Deposit date: | 2015-04-01 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Structural basis of enzymatic benzene ring reduction.
Nat.Chem.Biol., 11, 2015
|
|
3VHD
 
 | | Hsp90 alpha N-terminal domain in complex with a macrocyclic inhibitor, CH5164840 | | Descriptor: | 4-amino-18,20-dimethyl-7-thia-3,5,11,15-tetraazatricyclo[15.3.1.1(2,6)]docosa-1(20),2,4,6(22),17(21),18-hexaene-10,16-dione, Heat shock protein HSP 90-alpha | | Authors: | Fukami, T.A, Ono, N. | | Deposit date: | 2011-08-24 | | Release date: | 2012-07-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Design and synthesis of novel macrocyclic 2-amino-6-arylpyrimidine Hsp90 inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4FTQ
 
 | | Crystal Structure of the CHK1 | | Descriptor: | 5-{7-ethyl-6-[(3S)-tetrahydrofuran-3-yloxy]-2,4-dihydroindeno[1,2-c]pyrazol-3-yl}pyridine-2-carbonitrile, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
6R6D
 
 | | [Ru(TAP)2(11,12-CN2-dppz)]2+ bound to d(TCGGCGCCGA)2 | | Descriptor: | BARIUM ION, DNA (5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3'), Ruthenium (bis-(tetraazaphenanthrene)) (11,12-dicyano-dipyridophenazine), ... | | Authors: | McQuaid, K.T, Hall, J.P, Cardin, C.J. | | Deposit date: | 2019-03-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | X-ray Crystal Structures Show DNA Stacking Advantage of Terminal Nitrile Substitution in Ru-dppz Complexes.
Chemistry, 24, 2018
|
|
4ZAW
 
 | | Structure of UbiX in complex with reduced prenylated FMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, PHOSPHATE ION, Probable aromatic acid decarboxylase, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-14 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis.
Nature, 522, 2015
|
|
4PJV
 
 | | Structure of PARP2 catalytic domain bound to inhibitor BMN 673 | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Aoyagi-Scharber, M, Gardberg, A.S, Edwards, T.L. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the inhibition of poly(ADP-ribose) polymerases 1 and 2 by BMN 673, a potent inhibitor derived from dihydropyridophthalazinone.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4FVY
 
 | | Structure of rat nNOS heme domain in complex with N(omega)-hydroxy- N(omega)-methyl-L-arginine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2012-06-29 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Methylated N(omega)-Hydroxy-l-arginine Analogues as Mechanistic Probes for the Second Step of the Nitric Oxide Synthase-Catalyzed Reaction
Biochemistry, 52, 2013
|
|
4ZGA
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | (11aS)-6-(4-fluorobenzyl)-5,6,11,11a-tetrahydro-1H-imidazo[1',5':1,6]pyrido[3,4-b]indole-1,3(2H)-dione, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
4P6E
 
 | | Crystal Structure of Human Cathepsin S Bound to a Non-covalent Inhibitor | | Descriptor: | Cathepsin S, N-[(8R)-8-(benzoylamino)-5,6,7,8-tetrahydronaphthalen-2-yl]-4-methylpiperazine-1-carboxamide, SULFATE ION | | Authors: | Wang, Y, Jadhav, P.K, Deng, G.G. | | Deposit date: | 2014-03-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cathepsin S Inhibitor LY3000328 for the Treatment of Abdominal Aortic Aneurysm.
Acs Med.Chem.Lett., 5, 2014
|
|
4PNL
 
 | | Crystal structure of TNKS-2 in complex with DR2313. | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3,5,7,8-tetrahydro-4H-thiopyrano[4,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PCE
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with compound B13 | | Descriptor: | 1,2-ETHANEDIOL, 1-benzyl-2-ethyl-1,5,6,7-tetrahydro-4H-indol-4-one, Bromodomain-containing protein 4 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-04-15 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.293 Å) | | Cite: | Discovery of BRD4 bromodomain inhibitors by fragment-based high-throughput docking.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4PDK
 
 | | FadR, Fatty Acid Responsive Transcription Factor from Vibrio cholerae, in Complex with oleoyl-CoA | | Descriptor: | Fatty acid metabolism regulator protein, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Shi, W, Kull, F.J. | | Deposit date: | 2014-04-19 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 40-residue insertion in Vibrio cholerae FadR facilitates binding of an additional fatty acyl-CoA ligand.
Nat Commun, 6, 2015
|
|
3EJ8
 
 | | Structure of double mutant of human iNOS oxygenase domain with bound immidazole | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, HEME C, IMIDAZOLE, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-09-17 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
6A9O
 
 | | Rational discovery of a SOD1 tryptophan oxidation inhibitor with therapeutic potential for amyotrophic lateral sclerosis | | Descriptor: | 2'-[(6-oxo-5,6-dihydrophenanthridin-3-yl)carbamoyl][1,1'-biphenyl]-2-carboxylic acid, DIMETHYL SULFOXIDE, Dihydrogen tetrasulfide, ... | | Authors: | Manjula, R, Padmanabhan, B. | | Deposit date: | 2018-07-14 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational discovery of a SOD1 tryptophan oxidation inhibitor with therapeutic potential for amyotrophic lateral sclerosis.
J.Biomol.Struct.Dyn., 37, 2019
|
|
3VMT
 
 | | Crystal structure of Staphylococcus aureus membrane-bound transglycosylase in complex with a Lipid II analog | | Descriptor: | MAGNESIUM ION, Monofunctional glycosyltransferase, [(2R,3R,4R,5S,6R)-4-[(2R)-1-[[(2S)-1-[2-[2-[2-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]ethoxy]ethoxy]ethylamino]-1-oxidanylidene-propan-2-yl]amino]-1-oxidanylidene-propan-2-yl]oxy-3-acetamido-5-[(2S,3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-(hydroxymethyl)oxan-2-yl] [oxidanyl(3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaenoxy)phosphoryl] hydrogen phosphate | | Authors: | Huang, C.Y, Shih, H.W, Lin, L.Y, Tien, Y.W, Cheng, T.J.R, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FVX
 
 | | Structure of rat nNOS heme domain in complex with N(omega)-ethoxy-L-arginine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, brain, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2012-06-29 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Methylated N(omega)-Hydroxy-l-arginine Analogues as Mechanistic Probes for the Second Step of the Nitric Oxide Synthase-Catalyzed Reaction
Biochemistry, 52, 2013
|
|
