7SZ8
 
 | |
7QNY
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with COVOX-58 and COVOX-158 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7T3S
 
 | |
7TB0
 
 | | E. faecium MurAA in complex with fosfomycin and UNAG | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Zhou, Y, Shamoo, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enolpyruvate transferase MurAA A149E , identified during adaptation of Enterococcus faecium to daptomycin, increases stability of MurAA-MurG interaction.
J.Biol.Chem., 299, 2023
|
|
8EOG
 
 | | Structure of the human L-type voltage-gated calcium channel Cav1.2 complexed with L-leucine | | Descriptor: | (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(dodecanoyloxy)propyl dodecanoate, (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(dodecanoyloxy)propyl heptadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Mondal, A, Abderemane-Ali, F, Minor, D.L. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | EMC chaperone-Ca V structure reveals an ion channel assembly intermediate.
Nature, 619, 2023
|
|
7QBU
 
 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-methyl-5'-thioadenosine bound. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CO-METHYLCOBALAMIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7QBT
 
 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-methyl-5'-thioadenosine bound. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CO-METHYLCOBALAMIN, FE (III) ION, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7TAK
 
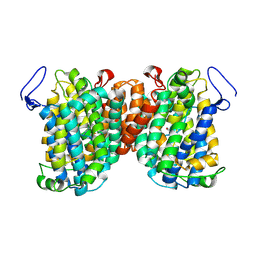 | | Structure of a NAT transporter | | Descriptor: | GUANINE, Putative membrane protein PurT | | Authors: | Weng, J, Zhou, X, Ren, Z, Chen, K, Zhou, M. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.79828 Å) | | Cite: | Insight into the mechanism of H + -coupled nucleobase transport.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2BII
 
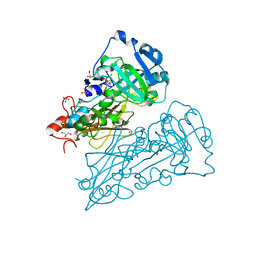 | | crystal structure of nitrate-reducing fragment of assimilatory nitrate reductase from Pichia angusta | | Descriptor: | (MOLYBDOPTERIN-S,S)-DIOXO-THIO-MOLYBDENUM(IV), GLYCEROL, NITRATE REDUCTASE [NADPH], ... | | Authors: | Fischer, K, Barbier, G, Hecht, H.-J, Mendel, R.R, Campbell, W.H, Schwarz, G. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Eukaryotic Nitrate Reduction: Crystal Structures of the Nitrate Reductase Active Site
Plant Cell, 17, 2005
|
|
7T8D
 
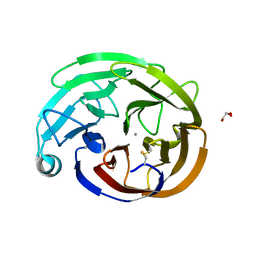 | | Myocilin OLF mutant V449I | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-12-16 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7QBS
 
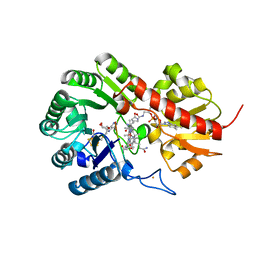 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, S-adenosyl-L-methionine, and peptide bound. | | Descriptor: | CO-METHYLCOBALAMIN, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Bernardo-Garcia, N, Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7QBV
 
 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, and S-adenosyl-L-homocysteine bound. | | Descriptor: | CO-METHYLCOBALAMIN, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
8EQG
 
 | |
8EKV
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGCGGATGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*CP*GP*GP*AP*TP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*AP*TP*CP*CP*GP*CP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-09-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8ENG
 
 | |
2BT3
 
 | | AGAO in complex with Ruthenium-C4-wire at 1.73 angstroms | | Descriptor: | BIS[1H,1'H-2,2'-BIPYRIDINATO(2-)-KAPPA~2~N~1~,N~1'~]{3-[4-(1,10-DIHYDRO-1,10-PHENANTHROLIN-4-YL-KAPPA~2~N~1~,N~10~)BUTOXY]-N,N-DIMETHYLANILINATO(2-)}RUTHENIUM, COPPER (II) ION, GLYCEROL, ... | | Authors: | Langley, D.B, Duff, A.P, Freeman, H.C, Guss, J.M, Juda, G.A, Dooley, D.M, Contakes, S.M, Halpern-Manners, N.W, Dunn, A.R, Gray, H.B. | | Deposit date: | 2005-05-26 | | Release date: | 2005-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Reversible Inhibition of Copper Amine Oxidase Activity by Channel-Blocking Ruthenium(II) and Rhenium(I) Molecular Wires.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
8EK3
 
 | |
8F3T
 
 | | Crystal structure of Penicillin Binding Protein 5 (PBP5) T485M T499I V629E variant apo form from Enterococcus faecium | | Descriptor: | Penicillin binding protein 5, SODIUM ION, SULFATE ION | | Authors: | D'Andrea, E.D, Choy, M.S, Schoenle, M.V, Page, R, Peti, W. | | Deposit date: | 2022-11-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The Molecular Basis for Resistance of E. faecium PBP5 to beta-lactam Antibiotics
Nat Commun, 2023
|
|
8EKZ
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGGAGAAGTAGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*GP*AP*GP*AP*AP*GP*TP*AP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*AP*CP*TP*TP*CP*TP*CP*CP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-09-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8EMD
 
 | |
8EO4
 
 | |
8EQK
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAACCGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-10-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8EQL
 
 | |
8JTO
 
 | | Outer membrane porin of Burkholderia pseudomallei (BpsOmp38) in complex with ceftazidime | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane porin (Fragment), SODIUM ION | | Authors: | Bunkum, P, Aunkham, A, Bert van den, B, Robinson, R.C, Suginta, W. | | Deposit date: | 2023-06-22 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and function of outer membrane protein from Burkholderia pseudomallei (BpsOmp38)
To be published
|
|
8ELM
 
 | | Apo human biliverdin reductase beta (293K) | | Descriptor: | Flavin reductase (NADPH), SODIUM ION | | Authors: | McLeod, M.J, Eisenmesser, E.Z, Lee, E, Thorne, R.E. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identifying structural and dynamic changes during the Biliverdin Reductase B catalytic cycle.
Front Mol Biosci, 10, 2023
|
|
