6KGH
 
 | | Crystal structure of Penicillin binding protein 3 (PBP3) from Mycobacterium tuerculosis (apo-form) | | Descriptor: | COBALT (II) ION, Penicillin-binding protein PbpB, SODIUM ION | | Authors: | Lu, Z.K, Zhang, A.L, Liu, X, Guddat, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2019-07-11 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | Structures ofMycobacterium tuberculosisPenicillin-Binding Protein 3 in Complex with Fivebeta-Lactam Antibiotics Reveal Mechanism of Inactivation.
Mol.Pharmacol., 97, 2020
|
|
1QMF
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) ACYL-ENZYME COMPLEX | | Descriptor: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, CEFUROXIME (OCT-3-ENE FORM), PENICILLIN-BINDING PROTEIN 2X | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Penicillin Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance
J.Mol.Biol., 299, 2000
|
|
3UN7
 
 | |
2WAF
 
 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN R6) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 2B | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
2JE5
 
 | | STRUCTURAL AND MECHANISTIC BASIS OF PENICILLIN BINDING PROTEIN INHIBITION BY LACTIVICINS | | Descriptor: | (2E)-2-{[(2S)-2-(ACETYLAMINO)-2-CARBOXYETHOXY]IMINO}PENTANEDIOIC ACID, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Macheboeuf, P, Fisher, D.S, Brown, T.J, Zervosen, A, Luxen, A, Joris, B, Dessen, A, Schofield, C.J. | | Deposit date: | 2007-01-15 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Basis of Penicillin-Binding Protein Inhibition by Lactivicins
Nat.Chem.Biol., 3, 2007
|
|
2JCH
 
 | | Structural and mechanistic basis of penicillin binding protein inhibition by lactivicins | | Descriptor: | (2E)-2-({(2S)-2-CARBOXY-2-[(PHENOXYACETYL)AMINO]ETHOXY}IMINO)PENTANEDIOIC ACID, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Macheboeuf, P, Fisher, D.S, Brown, T.J, Zervosen, A, Luxen, A, Joris, B, Dessen, A, Schofield, C.J. | | Deposit date: | 2006-12-23 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mechanistic Basis of Penicillin-Binding Protein Inhibition by Lactivicins
Nat.Chem.Biol., 3, 2007
|
|
3A3F
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae,complexed with novel beta-lactam (FMZ) | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxo-1-({(2R)-2-[(2-oxoimidazolidin-1-yl)amino]-2-phenylacetyl}amino)ethyl]-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | Authors: | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2009-06-12 | | Release date: | 2009-12-22 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
3A3I
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Haemophilus influenzae, complexed with ampicillin (AIX) | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 4 | | Authors: | Kawai, F, Roper, D.I, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2009-06-12 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae
J.Mol.Biol., 396, 2010
|
|
8C5O
 
 | |
8C5B
 
 | |
4K91
 
 | | Crystal structure of Penicillin-Binding Protein 5 (PBP5) from Pseudomonas aeruginosa in apo state | | Descriptor: | D-ala-D-ala-carboxypeptidase, SUCCINIC ACID | | Authors: | Smith, J, Toth, M, Vakulenko, S, Mobashery, S, Chen, Y. | | Deposit date: | 2013-04-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the role of Pseudomonas aeruginosa penicillin-binding protein 5 in beta-lactam resistance.
Antimicrob.Agents Chemother., 57, 2013
|
|
6I1G
 
 | |
6HZH
 
 | |
8U55
 
 | |
3MZF
 
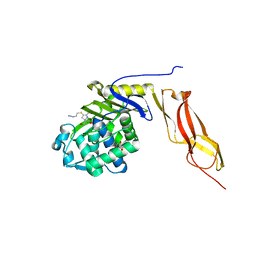 | | Structure of penicillin-binding protein 5 from E. coli: imipenem acyl-enzyme complex | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, D-alanyl-D-alanine carboxypeptidase dacA, GLYCEROL | | Authors: | Nicola, G, Tomberg, J, Pratt, R.F, Nicholas, R.A, Davies, C. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of covalent complexes of beta-lactam antibiotics with Escherichia coli penicillin-binding protein 5: toward an understanding of antibiotic specificity
Biochemistry, 49, 2010
|
|
3MZD
 
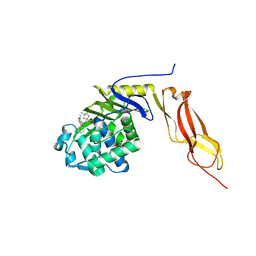 | | Structure of penicillin-binding protein 5 from E. coli: cloxacillin acyl-enzyme complex | | Descriptor: | (2R,4S)-2-[(1S)-1-({[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl}amino)-2-oxoethyl]-5,5-dimethyl-1,3-thiazolid ine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase dacA, GLYCEROL | | Authors: | Nicola, G, Tomberg, J, Pratt, R.F, Nicholas, R.A, Davies, C. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of covalent complexes of beta-lactam antibiotics with Escherichia coli penicillin-binding protein 5: toward an understanding of antibiotic specificity
Biochemistry, 49, 2010
|
|
3MZE
 
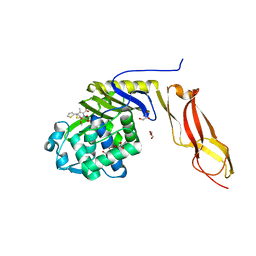 | | Structure of penicillin-binding protein 5 from E.coli: cefoxitin acyl-enzyme complex | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, D-alanyl-D-alanine carboxypeptidase dacA, GLYCEROL | | Authors: | Nicola, G, Tomberg, J, Pratt, R.F, Nicholas, R.A, Davies, C. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of covalent complexes of beta-lactam antibiotics with Escherichia coli penicillin-binding protein 5: toward an understanding of antibiotic specificity
Biochemistry, 49, 2010
|
|
3LO7
 
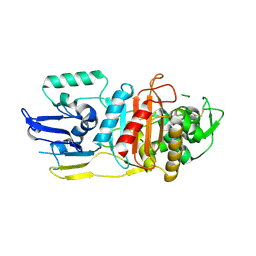 | |
3IT9
 
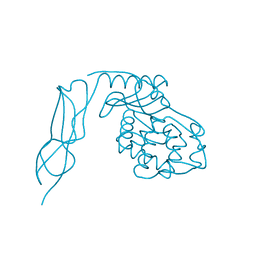 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in apo state | | Descriptor: | D-alanyl-D-alanine carboxypeptidase dacC, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
3ITB
 
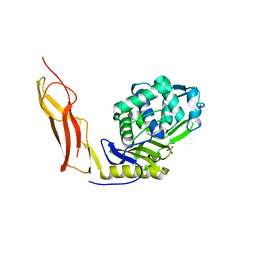 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in complex with a substrate fragment | | Descriptor: | D-alanyl-D-alanine carboxypeptidase DacC, Peptidoglycan substrate (AMV)A(FGA)K(DAL)(DAL), SULFATE ION, ... | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
3ITA
 
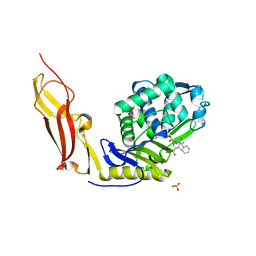 | | Crystal structure of Penicillin-Binding Protein 6 (PBP6) from E. coli in acyl-enzyme complex with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, (2S,5R,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, D-alanyl-D-alanine carboxypeptidase dacC, ... | | Authors: | Chen, Y, Zhang, W, Shi, Q, Hesek, D, Lee, M, Mobashery, S, Shoichet, B.K. | | Deposit date: | 2009-08-27 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of penicillin-binding protein 6 from Escherichia coli.
J.Am.Chem.Soc., 131, 2009
|
|
1KLX
 
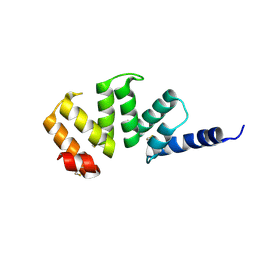 | |
6R40
 
 | |
5FGZ
 
 | | E. coli PBP1b in complex with FPI-1465 | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B, [[(3~{R},6~{S})-1-methanoyl-6-[[(3~{S})-pyrrolidin-3-yl]oxycarbamoyl]piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-12-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
6R3X
 
 | |
