2WZI
 
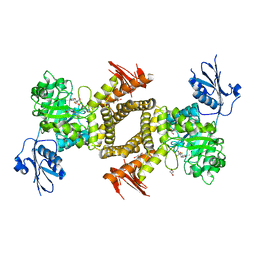 | | BtGH84 D243N in complex with 5F-oxazoline | | Descriptor: | (3AS,5S,6S,7R,7AR)-5-FLUORO-5-(HYDROXYMETHYL)-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]OXAZOLE-6,7-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Visualizing the Reaction Coordinate of an O-Glcnac Hydrolase
J.Am.Chem.Soc., 132, 2010
|
|
3ZJ0
 
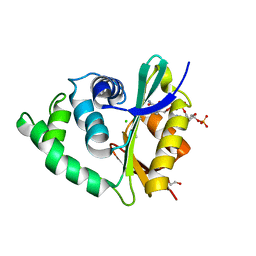 | | The human O-GlcNAcase C-terminal domain is a pseudo histone acetyltransferase | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, ACETYLTRANSFERASE, ... | | Authors: | Rao, F.V, Schuettelkopf, A.W, Dorfmueller, H.C, Ferenbach, A.T, Navratilova, I, van Aalten, D.M.F. | | Deposit date: | 2013-01-15 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Bacterial Putative Acetyltransferase Defines the Fold of the Human O-Glcnacase C-Terminal Domain.
Open Biol., 3, 2013
|
|
2XM2
 
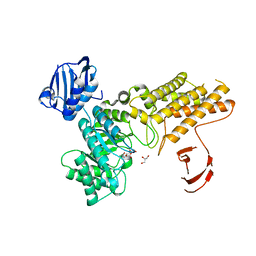 | | BtGH84 in complex with LOGNAc | | Descriptor: | GLYCEROL, N-acetylglucosaminono-1,5-lactone (Z)-oxime, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2010-07-22 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibition of a Bacterial O-Glcnacase Homologue by Lactone and Lactam Derivatives: Structural, Kinetic and Thermodynamic Analyses.
Amino Acids, 40, 2011
|
|
2XJ7
 
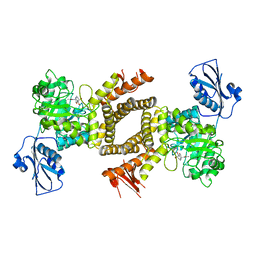 | | BtGH84 in complex with 6-acetamido-6-deoxy-castanospermine | | Descriptor: | 6-ACETAMIDO-6-DEOXY-CASTANOSPERMINE, CALCIUM ION, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of O-Glcnacase Using a Potent and Cell-Permeable Inhibitor Does not Induce Insulin Resistance in 3T3-L1 Adipocytes.
Chem.Biol., 17, 2010
|
|
2XM1
 
 | | BtGH84 in complex with N-acetyl gluconolactam | | Descriptor: | GLYCEROL, N-ACETYL GLUCONOLACTAM, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2010-07-22 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of a Bacterial O-Glcnacase Homologue by Lactone and Lactam Derivatives: Structural, Kinetic and Thermodynamic Analyses.
Amino Acids, 40, 2011
|
|
4KRV
 
 | | Crystal structure of catalytic domain of bovine beta1,4-galactosyltransferase mutant M344H-GalT1 complex with 6-sulfo-GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, 6-AMINOHEXYL-URIDINE-C1,5'-DIPHOSPHATE, Beta-1,4-galactosyltransferase 1, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2013-05-17 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Investigations on beta 1,4-galactosyltransferase I using 6-sulfo-GlcNAc as an acceptor sugar substrate.
Glycoconj J, 30, 2013
|
|
6R45
 
 | | Crystal structure of eukaryotic O-GlcNAcase HAT-like domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, O-GlcNAcase | | Authors: | Raimi, O.G, Gorelik, A, Hopkins-Navratilova, I, Aristotelous, T, Ferenbach, A, vanAalten, D. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Crystal structure of eukaryotic O-GlcNAcase HAT-like domain
To Be Published
|
|
2J47
 
 | | Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with a imidazole-pugnac hybrid inhibitor | | Descriptor: | (5R,6R,7R,8S)-8-(ACETYLAMINO)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-N-PHENYL-1,5,6,7,8,8A-HEXAHYDROIMIDAZO[1,2-A]PYRIDINE-2-CARBOXAMIDE, GLUCOSAMINIDASE, GLYCEROL | | Authors: | Dennis, R.J, Davies, G.J. | | Deposit date: | 2006-08-25 | | Release date: | 2006-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Inhibition of O-Glcnacase by a Gluco-Configured Nagstatin and a Pugnac-Imidazole Hybrid Inhibitor
Chem. Commun., 42, 2006
|
|
2JIW
 
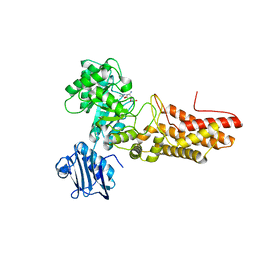 | | Bacteroides thetaiotaomicron GH84 O-GlcNAcase in complex with 2- Acetylamino-2-deoxy-1-epivalienamine | | Descriptor: | N-[(1S,2R,5R,6R)-2-AMINO-5,6-DIHYDROXY-4-(HYDROXYMETHYL)CYCLOHEX-3-EN-1-YL]ACETAMIDE, O-GLCNACASE BT_4395 | | Authors: | Dennis, R.J, Davies, G.J. | | Deposit date: | 2007-07-02 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A 1-Acetamido Derivative of 6-Epi-Valienamine: An Inhibitor of a Diverse Group of Beta-N-Acetylglucosaminidases.
Org.Biomol.Chem., 5, 2007
|
|
5UHO
 
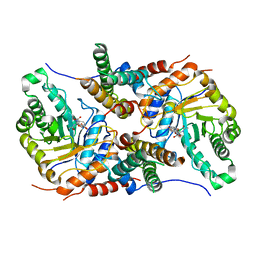 | |
5UHL
 
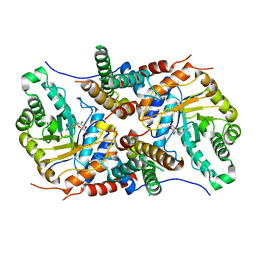 | | Crystal structure of the core catalytic domain of human O-GlcNAcase complexed with Thiamet G | | Descriptor: | (2Z,3aR,5R,6S,7R,7aR)-2-(ethylimino)-5-(hydroxymethyl)hexahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | Authors: | Klein, D.J, Elsen, N.L. | | Deposit date: | 2017-01-11 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of the core catalytic domain of human O-GlcNAcase and molecular basis of activity and inhibition
To Be Published
|
|
5UHK
 
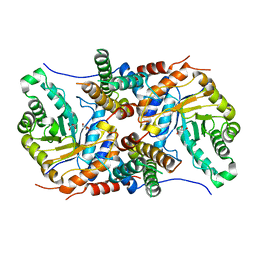 | |
5UHP
 
 | |
2W4X
 
 | | BtGH84 in complex with STZ | | Descriptor: | CALCIUM ION, GLYCEROL, O-GLCNACASE BT_4395, ... | | Authors: | He, Y, Bubb, A, Martinez-Fleites, C, Davies, G.J. | | Deposit date: | 2008-12-02 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Insight Into the Mechanism of Streptozotocin Inhibition of O-Glcnacase.
Carbohydr.Res., 344, 2009
|
|
6BZD
 
 | |
4XIF
 
 | | Human OGT in complex with UDP-5S-GlcNAc and substrate peptide (keratin-7) | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Keratin, type II cytoskeletal 7, ... | | Authors: | Schimpl, M, van Aalten, D.M.F. | | Deposit date: | 2015-01-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XI9
 
 | | Human OGT in complex with UDP-5S-GlcNAc and substrate peptide (RBL2) | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Retinoblastoma-like protein 2, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Schimpl, M, van Aalten, D.M.F. | | Deposit date: | 2015-01-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5C1D
 
 | | Human OGT in complex with UDP-5S-GlcNAc and substrate peptide (RB2L) | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, PHOSPHATE ION, Retinoblastoma-like protein 2, ... | | Authors: | Schimpl, M, Rafie, K, van Aalten, D.M.F. | | Deposit date: | 2015-06-13 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8CM9
 
 | | Structure of human O-GlcNAc transferase in complex with UDP and tP11 | | Descriptor: | PHE-MET-PRO-LYS-TYR-SER-ILE, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-5'-DIPHOSPHATE | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2023-02-18 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phage display uncovers a sequence motif that drives polypeptide binding to a conserved regulatory exosite of O-GlcNAc transferase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6Q4M
 
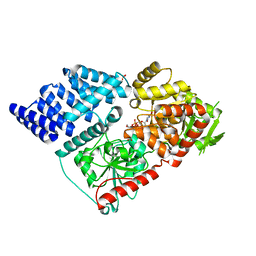 | | Crystal structure of the O-GlcNAc transferase Asn648Tyr mutation | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, PHOSPHATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Gundogdu, M, van Aalten, D.M.F. | | Deposit date: | 2018-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the O-GlcNAc transferase Asn648Tyr mutation
To Be Published
|
|
5LVV
 
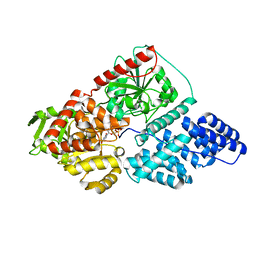 | | Human OGT in complex with UDP and fused substrate peptide (Tab1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit,UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-5'-DIPHOSPHATE | | Authors: | Raimi, O. | | Deposit date: | 2016-09-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats.
Open Biol, 7, 2017
|
|
5LWV
 
 | | Human OGT in complex with UDP and fused substrate peptide (HCF1) | | Descriptor: | GLYCEROL, Host cell factor 1,UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, PHOSPHATE ION, ... | | Authors: | Raimi, O, Rafie, K, Kapuria, V, Herr, W, van Aalten, D. | | Deposit date: | 2016-09-19 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats.
Open Biol, 7, 2017
|
|
6MA2
 
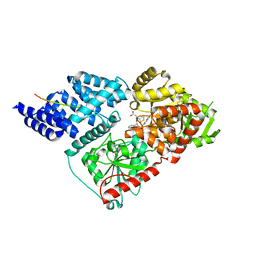 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor ent-1a | | Descriptor: | Host Cell Factor 1 peptide, N-[(2S)-2-(2-methoxyphenyl)-2-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
6MA3
 
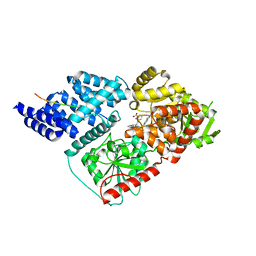 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 2a | | Descriptor: | 4-{2-[(1R)-2-{(carboxymethyl)[(thiophen-2-yl)methyl]amino}-2-oxo-1-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}ethyl]phenoxy}butanoic acid, Host Cell Factor 1 peptide, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
6MA4
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 3a | | Descriptor: | 5-{2-[(1R)-2-{(carboxymethyl)[(thiophen-2-yl)methyl]amino}-2-oxo-1-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}ethyl]phenoxy}pentanoic acid, Host Cell Factor 1 peptide, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
