5J13
 
 | |
5IWC
 
 | | Mycobacterium tuberculosis CysM in complex with the Urea-scaffold inhibitor 3 [4-(3-([1,1'-Biphenyl]-3-yl)ureido)-2-hydroxybenzoic acid] | | Descriptor: | 4-{[([1,1'-biphenyl]-3-yl)carbamoyl]amino}-2-hydroxybenzoic acid, O-phosphoserine sulfhydrylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schnell, R, Maric, S, Lindqvist, Y, Schneider, G. | | Deposit date: | 2016-03-22 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis.
J.Med.Chem., 59, 2016
|
|
6KSP
 
 | |
6KSS
 
 | |
5NHY
 
 | | BAY-707 in complex with MTH1 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, SULFATE ION, ... | | Authors: | Ellermann, M, Eheim, A, Giese, A, Bunse, S, Nowak-Reppel, K, Neuhaus, R, Weiske, J, Quanz, M, Glasauer, A, Meyer, H, Queisser, N, Irlbacher, H, Bader, B, Rahm, F, Viklund, J, Andersson, M, Ericsson, U, Ginman, T, Forsblom, R, Lindstrom, J, Silvander, C, Tresaugues, L, Gorjanacz, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel Class of Potent and Cellularly Active Inhibitors Devalidates MTH1 as Broad-Spectrum Cancer Target.
ACS Chem. Biol., 12, 2017
|
|
5LXJ
 
 | |
7B2P
 
 | | Cryo-EM structure of complement C4b in complex with nanobody B5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C4 alpha chain, ... | | Authors: | Oosterheert, W, De la O Becerra, K.I, Gros, P. | | Deposit date: | 2020-11-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Multifaceted Activities of Seven Nanobodies against Complement C4b.
J Immunol., 208, 2022
|
|
7B2M
 
 | | Cryo-EM structure of complement C4b in complex with nanobody E3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-C4b nanobody E3, Complement C4 alpha chain, ... | | Authors: | Oosterheert, W, De la O Becerra, K.I, van den Bos, R.M, Gros, P. | | Deposit date: | 2020-11-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Multifaceted Activities of Seven Nanobodies against Complement C4b.
J Immunol., 208, 2022
|
|
7B2Q
 
 | | Cryo-EM structure of complement C4b in complex with nanobody B12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-C4b nanobody B12, ... | | Authors: | Oosterheert, W, De la O Becerra, K.I, Gros, P. | | Deposit date: | 2020-11-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Multifaceted Activities of Seven Nanobodies against Complement C4b.
J Immunol., 208, 2022
|
|
7C1D
 
 | |
1KCA
 
 | |
1KN7
 
 | | Solution structure of the tandem inactivation domain (residues 1-75) of potassium channel RCK4 (Kv1.4) | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN KV1.4 | | Authors: | Wissmann, R, Bildl, W, Oliver, D, Beyermann, M, Kalbitzer, H.R, Bentrop, D, Fakler, B. | | Deposit date: | 2001-12-18 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Function of the "Tandem Inactivation Domain" of the Neuronal A-type
Potassium Channel Kv1.4
J.Biol.Chem., 278, 2003
|
|
5QCK
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(2~{S},3~{R})-1-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3-phenyl-pyrrolidin-2-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(2~{S},3~{R})-1-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3-phenyl-pyrrolidin-2-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
6Z2H
 
 | |
5QCM
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl ~{N}-[4-[[(1~{S})-2-[(~{E})-3-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]phenyl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
5QCL
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
5QCN
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-5-[(3~{S})-3-ethoxycarbonylpiperidin-1-yl]carbonyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-5-[(3~{S})-3-ethoxycarbonylpiperidin-1-yl]carbonyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
3GHG
 
 | | Crystal Structure of Human Fibrinogen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A knob, B knob, ... | | Authors: | Doolittle, R.F, Kollman, J.M, Sawaya, M.R, Pandi, L, Riley, M. | | Deposit date: | 2009-03-03 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human fibrinogen.
Biochemistry, 48, 2009
|
|
5YHJ
 
 | | Cytochrome P450EX alpha (CYP152N1) wild-type with myristic acid | | Descriptor: | Cytochrome P450, MYRISTIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Onoda, H, Shoji, O, Suzuki, K, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-09-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alpha-Oxidative Decarboxylation of Fatty Acids Catalysed by Cytochrome P450 Peroxygenases Yielding Shorter-Alkyl-Chain Fatty Acids
Catalysis Science And Technology, 2017
|
|
5ZSH
 
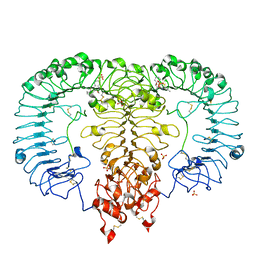 | | Crystal structure of monkey TLR7 in complex with CL075 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-propyl[1,3]thiazolo[4,5-c]quinolin-4-amine, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSL
 
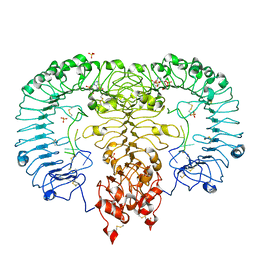 | | Crystal structure of monkey TLR7 in complex with GGUUGG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2S,3aR,4R,6R,6aR)-2-hydroxy-6-(hydroxymethyl)-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]-3,9-dihydro-6H-purin-6-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSD
 
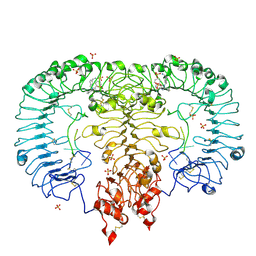 | | Crystal structure of monkey TLR7 in complex with IMDQ and GGUUGG | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSB
 
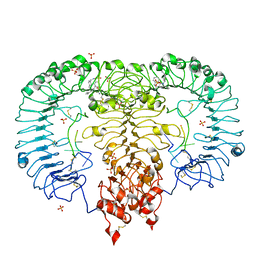 | | Crystal structure of monkey TLR7 in complex with IMDQ and AAUUAA | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]-2-butyl-imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSM
 
 | | Crystal structure of monkey TLR7 in complex with GGUCCC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2S,3aR,4R,6R,6aR)-2-hydroxy-6-(hydroxymethyl)-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]-3,9-dihydro-6H-purin-6-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
5ZSF
 
 | | Crystal structure of monkey TLR7 in complex with imiquimod | | Descriptor: | 1-(2-methylpropyl)imidazo[4,5-c]quinolin-4-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-04-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
