4ONU
 
 | |
4ORF
 
 | |
4OLL
 
 | |
2N0V
 
 | | Backbone 1H, Chemical Shift Assignments for Cn-APM1 | | Descriptor: | Antimicrobial peptide 1 | | Authors: | Santana, M.J, Oliveira, A.L, Queiroz Jr, L.K, Mandal, S.M, Matos, C.O, Dias, R.O, Franco, O.L, Liao, L.M. | | Deposit date: | 2015-03-17 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural insights into Cn-AMP1, a short disulfide-free multifunctional peptide from green coconut water.
Febs Lett., 589, 2015
|
|
5L0N
 
 | | PKG I's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with RP-cGMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-9-[(2R,4aR,6R,7R,7aS)-2,7-dihydroxy-2-sulfanylidenetetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-3,9-dihydro-6H-purin-6-one, CALCIUM ION, ... | | Authors: | Campbell, J.C, Sankaran, B, Kim, C.W. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.285 Å) | | Cite: | Structure of PKG I CNB-B bound to RP-cGMP
To Be Published
|
|
5JOC
 
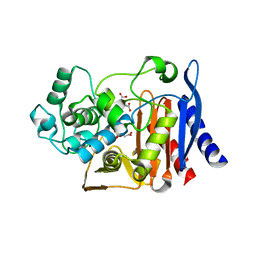 | | Crystal structure of the S61A mutant of AmpC BER | | Descriptor: | Beta-lactamase, CITRIC ACID | | Authors: | Na, J.H, An, Y.J, Cha, S.S. | | Deposit date: | 2016-05-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C beta-lactamases AmpC BER and CMY-10.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6UJA
 
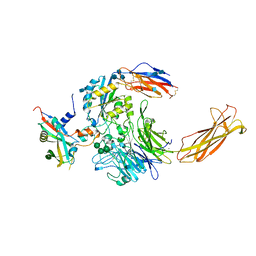 | | Integrin alpha-v beta-8 in complex with pro-TGF-beta1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Campbell, M.G, Cormier, A, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Reveals Integrin-Mediated TGF-beta Activation without Release from Latent TGF-beta.
Cell, 180, 2020
|
|
6I7L
 
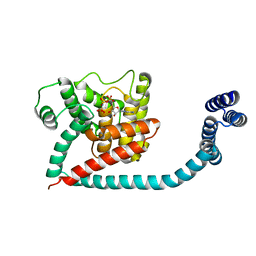 | | Crystal structure of monomeric FICD mutant L258D complexed with MgAMP-PNP | | Descriptor: | Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
4V2L
 
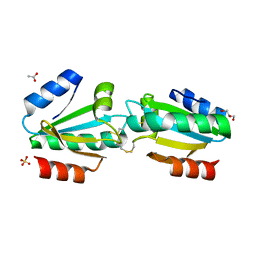 | |
4V2M
 
 | |
4V2N
 
 | |
4V7T
 
 | | Crystal structure of the E. coli ribosome bound to chloramphenicol. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.1942 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1L5I
 
 | | 30-CONFORMER NMR ENSEMBLE OF THE N-TERMINAL, DNA-BINDING DOMAIN OF THE REPLICATION INITIATION PROTEIN FROM A GEMINIVIRUS (TOMATO YELLOW LEAF CURL VIRUS-SARDINIA) | | Descriptor: | Rep protein | | Authors: | Campos-Olivas, R, Louis, J.M, Clerot, D, Gronenborn, B, Gronenborn, A.M. | | Deposit date: | 2002-03-07 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of a replication initiator unites diverse aspects of nucleic acid metabolism
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5XC4
 
 | | Crystal structure of GH45 endoglucanase EG27II at pH4.0, in complex with cellobiose | | Descriptor: | Endo-beta-1,4-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5XC9
 
 | | Crystal structure of GH45 endoglucanase EG27II at pH8.0, in complex with cellobiose | | Descriptor: | Endo-beta-1,4-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5XBU
 
 | | Crystal structure of GH45 endoglucanase EG27II in apo-form | | Descriptor: | Endo-beta-1,4-glucanase | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4V7W
 
 | | Structure of the Thermus thermophilus ribosome complexed with chloramphenicol. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5JIX
 
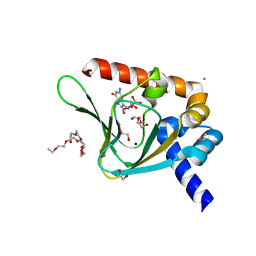 | | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-Br-cGMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-8-bromo-9-[(2R,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-1,9-dihydro-6H-purin-6-one, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Campbell, J.C, Kim, C.W. | | Deposit date: | 2016-04-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis of Analog Specificity in PKG I and II.
ACS Chem. Biol., 12, 2017
|
|
5JIZ
 
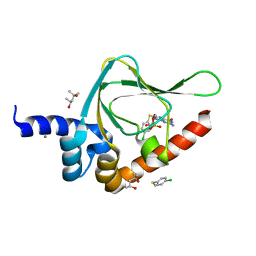 | | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-pCPT-cGMP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-amino-8-[(4-chlorophenyl)sulfanyl]-9-[(2S,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxotetrahydro-2H,4H-2lambda~5~-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-3,9-dihydro-6H-purin-6-one, CALCIUM ION, ... | | Authors: | Campbell, J.C, Kim, C.W. | | Deposit date: | 2016-04-22 | | Release date: | 2017-05-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | PKG II's Carboxyl Terminal Cyclic Nucleotide Binding Domain (CNB-B) in a complex with 8-pCPT-cGMP
To Be Published
|
|
7OCE
 
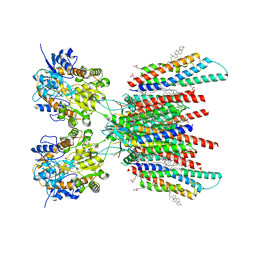 | | Resting state GluA1/A2 AMPA receptor in complex with TARP gamma 8 and CNIH2 (LBD-TMD) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, CHOLESTEROL, ... | | Authors: | Zhang, D, Watson, J.F, Matthews, P.M, Cais, O, Greger, I.H. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Gating and modulation of a hetero-octameric AMPA glutamate receptor.
Nature, 594, 2021
|
|
7OCF
 
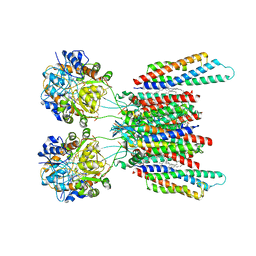 | | Active state GluA1/A2 AMPA receptor in complex with TARP gamma 8 and CNIH2 (LBD-TMD) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLOTHIAZIDE, GLUTAMIC ACID, ... | | Authors: | Zhang, D, Watson, J.F, Matthews, P.M, Cais, O, Greger, I.H. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Gating and modulation of a hetero-octameric AMPA glutamate receptor.
Nature, 594, 2021
|
|
5N1O
 
 | |
5N32
 
 | |
5N1K
 
 | |
7RZ8
 
 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
