5HU6
 
 | |
3VV8
 
 | | Crystal structure of beta secetase in complex with 2-amino-3-methyl-6-((1S,2R)-2-(3'-methylbiphenyl-4-yl)cyclopropyl)pyrimidin-4(3H)-one | | Descriptor: | 2-amino-3-methyl-6-[(1S,2R)-2-(3'-methylbiphenyl-4-yl)cyclopropyl]pyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL | | Authors: | Yonezawa, S, Yamamoto, T, Yamakawa, H, Muto, C, Hosono, M, Hattori, K, Higashino, K, Sakagami, M, Togame, H, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors: effect of a cyclopropane ring to induce an alternative binding mode.
J.Med.Chem., 55, 2012
|
|
5E7D
 
 | | Crystal Structure of the fifth bromodomain of human PB1 in complex with a hydroxyphenyl ligand | | Descriptor: | (2E)-3-(dimethylamino)-1-(2-hydroxyphenyl)prop-2-en-1-one, 1,2-ETHANEDIOL, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Owen, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-12 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of the fifth bromodomain of human PB1 in complex with a hydroxyphenyl ligand
To Be Published
|
|
2OQC
 
 | | Crystal Structure of Penicillin V acylase from Bacillus subtilis | | Descriptor: | Penicillin V acylase | | Authors: | Suresh, C.G, Rathinaswamy, P, Pundle, A.V, Prabhune, A.A, Sivaraman, H, Brannigan, J.A, Dodson, G.G. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Penicillin V acylase from Bacillus subtilis
to be published
|
|
3U1I
 
 | | Dengue virus protease covalently bound to a peptide | | Descriptor: | SULFATE ION, Serine protease NS3, Serine protease subunit NS2B, ... | | Authors: | Noble, C.G. | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand-bound structures of the dengue virus protease reveal the active conformation
J.Virol., 86, 2012
|
|
3AF1
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with GDP | | Descriptor: | CHLORIDE ION, CITRATE ANION, GLYCEROL, ... | | Authors: | Chetnani, B, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2010-02-19 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | M. tuberculosis pantothenate kinase: dual substrate specificity and unusual changes in ligand locations
J.Mol.Biol., 400, 2010
|
|
7BAQ
 
 | |
7BAR
 
 | |
3AF3
 
 | | Pantothenate kinase from Mycobacterium tuberculosis (MtPanK) in complex with GMPPCP and Pantothenate | | Descriptor: | GLYCEROL, PANTOTHENOIC ACID, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Chetnani, B, Kumar, P, Surolia, A, Vijayan, M. | | Deposit date: | 2010-02-22 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | M. tuberculosis pantothenate kinase: dual substrate specificity and unusual changes in ligand locations
J.Mol.Biol., 400, 2010
|
|
3JD0
 
 | | Glutamate dehydrogenase in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD4
 
 | | Glutamate dehydrogenase in complex with NADH and GTP, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3OBT
 
 | |
3WB5
 
 | | Crystal Structure of beta secetase in complex with (6S)-2-amino-3,6-dimethyl-6-[(1R,2R)-2-phenylcyclopropyl]-3,4,5,6-tetrahydropyrimidin-4-one | | Descriptor: | (6S)-2-amino-3,6-dimethyl-6-[(1R,2R)-2-phenylcyclopropyl]-5,6-dihydropyrimidin-4(3H)-one, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Yonezawa, S, Fujiwara, K, Yamamoto, T, Hattori, K, Yamakawa, H, Muto, C, Hosono, M, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2013-05-13 | | Release date: | 2013-10-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors III: Effective investigation of the binding mode by combinational use of X-ray analysis, isothermal titration calorimetry and theoretical calculations
Bioorg.Med.Chem., 21, 2013
|
|
2QBO
 
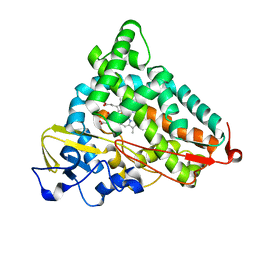 | | Crystal structure of the P450cam G248V mutant in the cyanide bound state | | Descriptor: | CAMPHOR, CYANIDE ION, Cytochrome P450-cam, ... | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.D, Schlichting, I. | | Deposit date: | 2007-06-18 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alteration of P450 Distal Pocket Solvent Leads to Impaired Proton Delivery and Changes in Heme Geometry.
Biochemistry, 46, 2007
|
|
3WVR
 
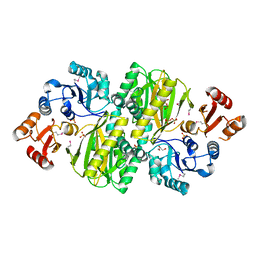 | | Structure of ATP grasp protein with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PGM1, ... | | Authors: | Matsui, T, Noike, M, Ooya, K, Sasaki, I, Hamano, Y, Maruyama, C, Ishikawa, J, Satoh, Y, Ito, H, Dairi, T, Morita, H. | | Deposit date: | 2014-06-04 | | Release date: | 2014-11-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin.
Nat.Chem.Biol., 11, 2015
|
|
1OSZ
 
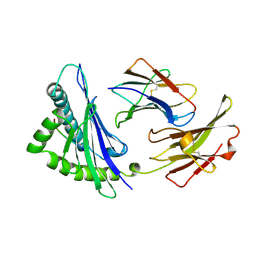 | | MHC CLASS I H-2KB HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND AN (L4V) MUTANT OF THE VESICULAR STOMATITIS VIRUS NUCLEOPROTEIN | | Descriptor: | BETA-2 MICROGLOBULIN, MHC CLASS I H-2KB HEAVY CHAIN, VESICULAR STOMATITIS VIRUS NUCLEOPROTEIN | | Authors: | Ghendler, Y, Teng, M.-K, Liu, J.-H, Witte, T, Liu, J, Kim, K.S, Kern, P, Chang, H.-C, Wang, J.-H, Reinherz, E.L. | | Deposit date: | 1998-06-18 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Differential thymic selection outcomes stimulated by focal structural alteration in peptide/major histocompatibility complex ligands.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
7F16
 
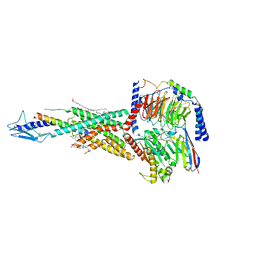 | | Cryo-EM structure of parathyroid hormone receptor type 2 in complex with a tuberoinfundibular peptide of 39 residues and G protein | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, X, Cheng, X, Zhao, L.H, Wang, Y, Ye, C, Zou, X, Dai, A, Cong, Z, Chen, J, Zhou, Q, Xia, T, Jiang, H, Xu, H.E, Yang, D, Wang, M.W. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular insights into differentiated ligand recognition of the human parathyroid hormone receptor 2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3VV7
 
 | | Crystal Structure of beta secetase in complex with 2-amino-6-((1S,2R)-2-(3'-methoxybiphenyl-3-yl)cyclopropyl)-3-methylpyrimidin-4(3H)-one | | Descriptor: | 2-amino-6-[(1S,2R)-2-(3'-methoxybiphenyl-3-yl)cyclopropyl]-3-methylpyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL, ... | | Authors: | Yonezawa, S, Yamamoto, T, Yamakawa, H, Muto, C, Hosono, M, Hattori, K, Higashino, K, Sakagami, M, Togame, H, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors: effect of a cyclopropane ring to induce an alternative binding mode.
J.Med.Chem., 55, 2012
|
|
4CPB
 
 | | CRYSTAL STRUCTURE OF LECA IN COMPLEX WITH A DIVALENT GALACTOSIDE AT 1. 57 ANGSTROM IN MAGNESIUM | | Descriptor: | (4S)-N-ethyl-4-{[N-methyl-3-(1-{2-[(4-sulfanylbenzoyl)amino]ethyl}-1H-1,2,3-triazol-4-yl)-L-alanyl]amino}-L-prolinamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Topin, J, Varrot, A, Imberty, A, Wissinger, N. | | Deposit date: | 2014-02-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A Leca Ligand Identified from a Galactoside-Conjugate Array Inhibits Host Cell Invasion by Pseudomonas Aeruginosa.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD3
 
 | | Glutamate dehydrogenase in complex with NADH and GTP, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3WVQ
 
 | | Structure of ATP grasp protein | | Descriptor: | GLYCEROL, PGM1, SULFATE ION | | Authors: | Matsui, T, Noike, M, Ooya, K, Sasaki, I, Hamano, Y, Maruyama, C, Ishikawa, J, Satoh, Y, Ito, H, Dairi, T, Morita, H. | | Deposit date: | 2014-06-04 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | A peptide ligase and the ribosome cooperate to synthesize the peptide pheganomycin.
Nat.Chem.Biol., 11, 2015
|
|
4CP9
 
 | | Crystal structure OF lecA lectin complexed with a divalent galactoside at 1.65 angstrom | | Descriptor: | (4S)-N-ethyl-4-{[N-methyl-3-(1-{2-[(4-sulfanylbenzoyl)amino]ethyl}-1H-1,2,3-triazol-4-yl)-L-alanyl]amino}-L-prolinamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Topin, J, Varrot, A, Imberty, A, Wissinger, N. | | Deposit date: | 2014-02-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Leca Ligand Identified from a Galactoside-Conjugate Array Inhibits Host Cell Invasion by Pseudomonas Aeruginosa.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3WB4
 
 | | Crystal Structure of beta secetase in complex with 2-amino-3,6-dimethyl-6-(2-phenylethyl)-3,4,5,6-tetrahydropyrimidin-4-one | | Descriptor: | (6R)-2-amino-3,6-dimethyl-6-(2-phenylethyl)-5,6-dihydropyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL, ... | | Authors: | Yonezawa, S, Fujiwara, K, Yamamoto, T, Hattori, K, Yamakawa, H, Muto, C, Hosono, M, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2013-05-13 | | Release date: | 2013-10-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors III: Effective investigation of the binding mode by combinational use of X-ray analysis, isothermal titration calorimetry and theoretical calculations
Bioorg.Med.Chem., 21, 2013
|
|
1TQQ
 
 | | Structure of TolC in complex with hexamminecobalt | | Descriptor: | COBALT HEXAMMINE(III), Outer membrane protein tolC | | Authors: | Higgins, M.K, Eswaran, J, Edwards, P.C, Schertler, G.F, Hughes, C, Koronakis, V. | | Deposit date: | 2004-06-18 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the ligand-blocked periplasmic entrance of the bacterial multidrug effllux protein TolC
J.Mol.Biol., 342, 2004
|
|
