3K9N
 
 | | Allosteric modulation of H-Ras GTPase | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Fetics, S, Young, M, Buhrman, G, Mattos, C. | | Deposit date: | 2009-10-16 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric modulation of Ras positions Q61 for a direct role in catalysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2L0S
 
 | | Solution Structure of Human Plasminogen Kringle 3 | | Descriptor: | Plasminogen | | Authors: | Christen, M.T, Frank, P, Schaller, J, Llinas, M. | | Deposit date: | 2010-07-15 | | Release date: | 2010-08-04 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Human plasminogen kringle 3: solution structure, functional insights, phylogenetic landscape.
Biochemistry, 49, 2010
|
|
2LKC
 
 | | Free B.st IF2-G2 | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Wienk, H, Tishchenko, E, Belardinelli, R, Tomaselli, S, Dongre, R, Spurio, R, Folkers, G.E, Gualerzi, C.O, Boelens, R. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Dynamics of Bacterial Translation Initiation Factor IF2.
J.Biol.Chem., 287, 2012
|
|
3JVW
 
 | | HIV-1 Protease Mutant G86A with symmetric inhibitor DMP323 | | Descriptor: | Gag-Pol polyprotein, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3JW2
 
 | | HIV-1 Protease Mutant G86S with DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
2I1P
 
 | | Solution structure of the twelfth cysteine-rich ligand-binding repeat in rat megalin | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 2 | | Authors: | Wolf, C.A, Dancea, F, Shi, M, Bade-Noskova, V, Rueterjans, H, Kerjaschki, D, Luecke, C. | | Deposit date: | 2006-08-14 | | Release date: | 2007-02-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the twelfth cysteine-rich ligand-binding repeat in rat megalin.
J.Biomol.Nmr, 37, 2007
|
|
2HXS
 
 | |
2I0D
 
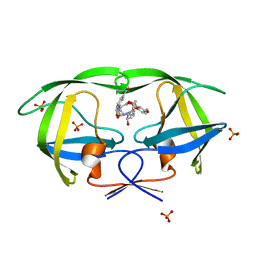 | | Crystal structure of AD-81 complexed with wild type HIV-1 protease | | Descriptor: | (5S)-3-(3-ACETYLPHENYL)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]-2-OXO-1,3-OXAZOLIDINE-5-CARBOXAMIDE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A, Ali, A, Reddy, K.K, Cao, H, Anjum, S.G, Rana, T.M. | | Deposit date: | 2006-08-10 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of HIV-1 Protease Inhibitors with Picomolar Affinities Incorporating N-Aryl-oxazolidinone-5-carboxamides as Novel P2 Ligands.
J.Med.Chem., 49, 2006
|
|
2I0H
 
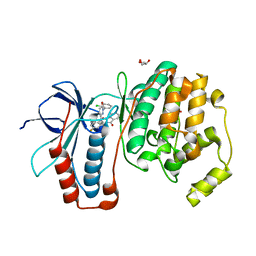 | | The structure of p38alpha in complex with an arylpyridazinone | | Descriptor: | 2-(3-{(2-CHLORO-4-FLUOROPHENYL)[1-(2-CHLOROPHENYL)-6-OXO-1,6-DIHYDROPYRIDAZIN-3-YL]AMINO}PROPYL)-1H-ISOINDOLE-1,3(2H)-DIONE, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Natarajan, S.R, Heller, S.T, Nam, K, Singh, S.B, Scapin, G, Patel, S, Thompson, J.E, Fitzgerald, C.E, O'Keefe, S.J. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p38 MAP Kinase Inhibitors Part 6: 2-Arylpyridazin-3-ones as templates for inhibitor design.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2I13
 
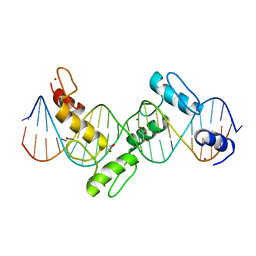 | | Aart, a six finger zinc finger designed to recognize ANN triplets | | Descriptor: | 5'-D(*CP*AP*GP*AP*TP*GP*TP*AP*GP*GP*GP*AP*AP*AP*AP*GP*CP*CP*CP*GP*GP*G)-3', 5'-D(*GP*CP*CP*CP*GP*GP*GP*CP*TP*TP*TP*TP*CP*CP*CP*TP*AP*CP*AP*TP*CP*T)-3', Aart, ... | | Authors: | Horton, N.C, Segal, D.J, Bhakta, M, Crotty, J.W, Barbas III, C.F. | | Deposit date: | 2006-08-12 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of Aart, a Designed Six-finger Zinc Finger Peptide, Bound to DNA.
J.Mol.Biol., 363, 2006
|
|
2I1U
 
 | |
3JUH
 
 | |
2I6F
 
 | | Receiver domain from Myxococcus xanthus social motility protein FrzS | | Descriptor: | CHLORIDE ION, Response regulator FrzS | | Authors: | Echols, N, Fraser, J, Merlie, J, Zusman, D, Alber, T. | | Deposit date: | 2006-08-28 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS.
Mol.Microbiol., 65, 2007
|
|
3JXI
 
 | |
2I6Z
 
 | |
2I8F
 
 | |
3K9L
 
 | | Allosteric modulation of H-Ras GTPase | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Fetics, S, Young, M, Buhrman, G, Mattos, C. | | Deposit date: | 2009-10-15 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric modulation of H-Ras GTPase
To be Published
|
|
3K2P
 
 | | HIV-1 Reverse Transcriptase Isolated RnaseH Domain with the Inhibitor beta-thujaplicinol Bound at the Active Site | | Descriptor: | 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, MANGANESE (II) ION, Reverse Transcriptase | | Authors: | Pauly, T.A, Himmel, D.M, Maegley, K, Arnold, E. | | Deposit date: | 2009-09-30 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site.
Structure, 17, 2009
|
|
3K3A
 
 | |
3K5F
 
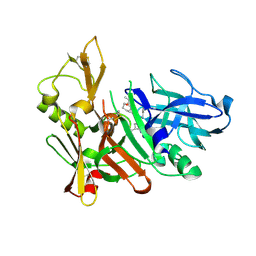 | | Human BACE-1 COMPLEX WITH AYH011 | | Descriptor: | (1R,3S)-3-[1-(acetylamino)-1-methylethyl]-N-[(1S,2S,4R)-1-benzyl-5-(butylamino)-2-hydroxy-4-methyl-5-oxopentyl]cyclohexanecarboxamide, Beta-secretase 1 | | Authors: | Rondeau, J.-M. | | Deposit date: | 2009-10-07 | | Release date: | 2010-05-05 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design and synthesis of novel P2/P3 modified, non-peptidic beta-secretase (BACE-1) inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2I5S
 
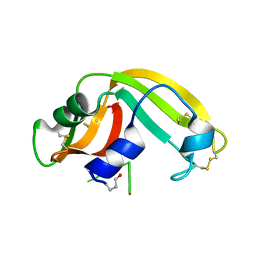 | | Crystal structure of onconase with bound nucleic acid | | Descriptor: | 5'-D(*A*(DU)P*GP*A)-3', P-30 protein | | Authors: | Bae, E, Lee, J.E, Raines, R.T, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for catalysis by onconase.
J.Mol.Biol., 375, 2008
|
|
3K8B
 
 | |
2I0A
 
 | | Crystal Structure of KB-19 complexed with wild type HIV-1 protease | | Descriptor: | (5S)-3-(4-ACETYLPHENYL)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]-2-OXO-1,3-OXAZOLIDINE-5-CARBOXAMIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A, Ali, A, Reddy, K.K, Cao, H, Anjum, S.G, Rana, T.M. | | Deposit date: | 2006-08-10 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of HIV-1 Protease Inhibitors with Picomolar Affinities Incorporating N-Aryl-oxazolidinone-5-carboxamides as Novel P2 Ligands.
J.Med.Chem., 49, 2006
|
|
2HZR
 
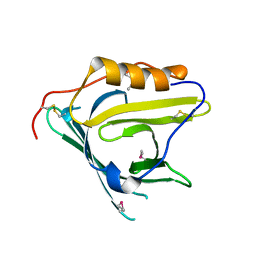 | | Crystal structure of human apolipoprotein D (ApoD) | | Descriptor: | Apolipoprotein D | | Authors: | Eichinger, A, Skerra, A. | | Deposit date: | 2006-08-09 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the dual ligand specificity and mode of high density lipoprotein association of apolipoprotein d.
J.Biol.Chem., 282, 2007
|
|
3JR8
 
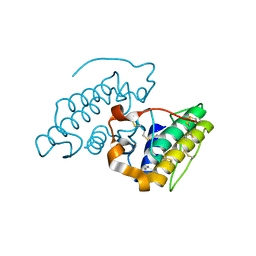 | | Crystal Structure of BthTX-II (Asp49-PLA2 from Bothrops jararacussu snake venom) with calcium ions | | Descriptor: | CALCIUM ION, Phospholipase A2 bothropstoxin-2 | | Authors: | Borges, R.J, dos Santos, J.I, Fontes, M.R.M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, functional, and bioinformatics studies reveal a new snake venom homologue phospholipase A2 class.
Proteins, 79, 2011
|
|
