9L6K
 
 | |
4EXP
 
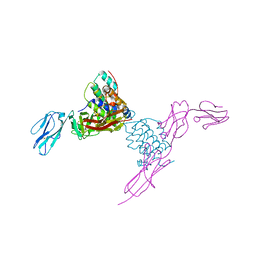 | | Structure of mouse Interleukin-34 in complex with mouse FMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-34, Macrophage colony-stimulating factor 1 receptor | | Authors: | Liu, H, Leo, C, Chen, X, Wong, B.R, Williams, L.T, Lin, H, He, X. | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1.
Biochim.Biophys.Acta, 1824, 2012
|
|
4Y87
 
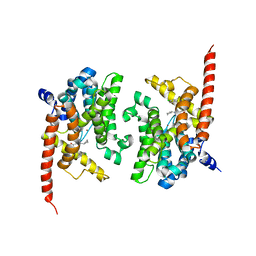 | | Crystal structure of phosphodiesterase 9 in complex with (R)-C33 (6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one) | | Descriptor: | 6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Huang, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
2MXA
 
 | | Solution structure of the NDH-1 complex subunit CupS from Thermosynechococcus elongatus | | Descriptor: | NDH-1 complex sensory subunit CupS | | Authors: | Korste, A, Wulfhorst, H, Ikegami, T, Nowaczyk, M.M, Stoll, R. | | Deposit date: | 2014-12-17 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the NDH-1 complex subunit CupS from Thermosynechococcus elongatus.
Biochim.Biophys.Acta, 1847, 2015
|
|
2N5L
 
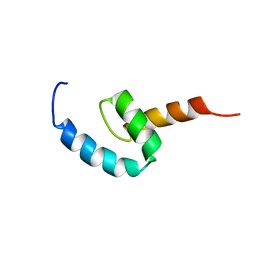 | | Regnase-1 C-terminal domain | | Descriptor: | Ribonuclease ZC3H12A | | Authors: | Yokogawa, M, Tsushima, T, Noda, N.N, Kumeta, H, Adachi, W, Enokizono, Y, Yamashita, K, Standley, D.M, Takeuchi, O, Akira, S, Inagaki, F. | | Deposit date: | 2015-07-18 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
9CGD
 
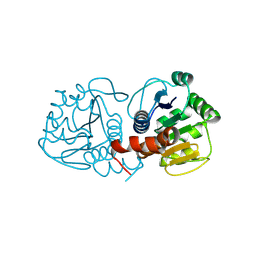 | | Human DJ-1, 10 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography, CrystFEL processed | | Descriptor: | Parkinson disease protein 7 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-28 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
4Y76
 
 | | Factor Xa complex with GTC000401 | | Descriptor: | CALCIUM ION, Coagulation factor X, N~2~-[(6-chloronaphthalen-2-yl)sulfonyl]-N~2~-{(3S)-1-[(2S)-1-(4-methyl-1,4-diazepan-1-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}glycinamide | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
9CFQ
 
 | | Human DJ-1, no mixing, pink beam time-resolved serial crystallography, CrystFEL processed | | Descriptor: | Parkinson disease protein 7 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-27 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CFY
 
 | | Human DJ-1, 15 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography | | Descriptor: | Parkinson disease protein 7 | | Authors: | Zielinski, K.A, Dolamore, C, Dalton, K, Meisburger, S.P, Smith, N, Termini, T, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-27 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CGG
 
 | | Human DJ-1, 30 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography, CrystFEL processed | | Descriptor: | Parkinson disease protein 7 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-28 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CG0
 
 | | Human DJ-1, 30 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography | | Descriptor: | Parkinson disease protein 7 | | Authors: | Zielinski, K.A, Dolamore, C, Dalton, K, Meisburger, S.P, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-28 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CEI
 
 | | Human DJ-1, no mixing, pink beam time-resolved serial crystallography | | Descriptor: | Parkinson disease protein 7 | | Authors: | Zielinski, K.A, Dolamore, C, Dalton, K, Meisburger, S.P, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-26 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CGE
 
 | | Human DJ-1, 15 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography, CrystFEL processed | | Descriptor: | Parkinson disease protein 7 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-28 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CFO
 
 | | Human DJ-1, 10 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography | | Descriptor: | Parkinson disease protein 7 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S.P, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-27 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9D50
 
 | | Structure of PAK1 in complex with compound 24 | | Descriptor: | 3-cyano-N-[3-({6-[(5-cyclopropyl-1,3-thiazol-2-yl)amino]pyrazin-2-yl}amino)bicyclo[1.1.1]pentan-1-yl]azetidine-3-carboxamide, Serine/threonine-protein kinase PAK 1 | | Authors: | Fontano, E, Suto, R.K, Olland, A.M. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 127, 2025
|
|
1TQJ
 
 | | Crystal structure of D-ribulose 5-phosphate 3-epimerase from Synechocystis to 1.6 angstrom resolution | | Descriptor: | Ribulose-phosphate 3-epimerase | | Authors: | Wise, E.L, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-06-17 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of D-ribulose 5-phosphate 3-epimerase from Synechocystis to 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3U7B
 
 | | A new crystal structure of a Fusarium oxysporum GH10 xylanase reveals the presence of an extended loop on top of the catalytic cleft | | Descriptor: | 1,2-ETHANEDIOL, ENDO-1,4-BETA-XYLANASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dimarogona, M, Topakas, E, Christakopoulos, P, Chrysina, E.D. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The structure of a GH10 xylanase from Fusarium oxysporum reveals the presence of an extended loop on top of the catalytic cleft.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
9CPH
 
 | | Structural basis of BAK sequestration by MCL-1 and consequences for apoptosis initiation | | Descriptor: | Bcl-2 homologous antagonist/killer, Induced myeloid leukemia cell differentiation protein Mcl-1, Synthetic antibody, ... | | Authors: | Uchikawa, E, Myasnikov, A, Dey, R, Moldoveanu, T. | | Deposit date: | 2024-07-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis of BAK sequestration by MCL-1 in apoptosis.
Mol.Cell, 85, 2025
|
|
4EU2
 
 | | Crystal structure of 20s proteasome with novel inhibitor K-7174 | | Descriptor: | 1,4-bis[(4E)-5-(3,4,5-trimethoxyphenyl)pent-4-en-1-yl]-1,4-diazepane, Proteasome component C1, Proteasome component C11, ... | | Authors: | Kikuchi, J, Shibayama, N, Yamada, S, Wada, T, Nobuyoshi, M, Izumi, T, Akutsu, M, Kano, Y, Ohki, M, Sugiyama, K, Park, S.-Y, Furukawa, Y. | | Deposit date: | 2012-04-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Homopiperazine derivatives as a novel class of proteasome inhibitors with a unique mode of proteasome binding.
Plos One, 8, 2013
|
|
1TOZ
 
 | | NMR structure of the human NOTCH-1 ligand binding region | | Descriptor: | Neurogenic locus notch homolog protein 1 | | Authors: | Hambleton, S, Valeyev, N.Y, Muranyi, A, Knott, V, Werner, J.M, Mcmichael, A.J, Handford, P.A, Downing, A.K. | | Deposit date: | 2004-06-15 | | Release date: | 2004-10-12 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural and functional properties of the human notch-1 ligand binding region
STRUCTURE, 12, 2004
|
|
4END
 
 | | Crystal structure of anti-HIV actinohivin in complex with alpha-1,2-mannobiose (P 2 21 21 form) | | Descriptor: | ACETONITRILE, Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-07-17 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peculiarity in crystal packing of anti-HIV lectin actinohivin in complex with alpha (1-2)mannobiose.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
9CF8
 
 | |
4YAB
 
 | | Crystal structure of TRIM24 PHD-bromodomain complexed with 1-methyl-5-(2-methyl-1 3-thiazol-4-yl)-2 3-dihydro-1H-indol-2-one (1) | | Descriptor: | 1-methyl-5-(2-methyl-1,3-thiazol-4-yl)-1,3-dihydro-2H-indol-2-one, SULFATE ION, Transcription intermediary factor 1-alpha, ... | | Authors: | Poncet-Montange, G, Palmer, W, Jones, P. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of IACS-9571, a Selective High-Affinity Dual TRIM24-BRPF1 Bromodomain Inhibitor.
J.Med.Chem., 59, 2016
|
|
1U57
 
 | | NMR structure of the (345-392)Gag sequence from HIV-1 | | Descriptor: | Gag polyprotein | | Authors: | Morellet, N, Druillennec, S, Lenoir, C, Bouaziz, S, Roques, B.P. | | Deposit date: | 2004-07-27 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Helical structure determined by NMR of the HIV-1 (345-392)Gag sequence, surrounding p2: Implications for particle assembly and RNA packaging
Protein Sci., 14, 2005
|
|
1TS5
 
 | | I140T MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
