4BUC
 
 | | CRYSTAL STRUCTURE OF MURD LIGASE FROM THERMOTOGA MARITIMA IN APO FORM | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Favini-Stabile, S, Contreras-Martel, C, Thielens, N, Dessen, A. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Mreb and Murg as Scaffolds for the Cytoplasmic Steps of Peptidoglycan Biosynthesis
Environ.Microbiol., 15, 2013
|
|
1COA
 
 | | THE EFFECT OF CAVITY CREATING MUTATIONS IN THE HYDROPHOBIC CORE OF CHYMOTRYPSIN INHIBITOR 2 | | Descriptor: | CHYMOTRYPSIN INHIBITOR 2 | | Authors: | Jackson, S.E, Moracci, M, Elmasry, N, Johnson, C.M, Fersht, A.R. | | Deposit date: | 1993-05-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effect of cavity-creating mutations in the hydrophobic core of chymotrypsin inhibitor 2.
Biochemistry, 32, 1993
|
|
1D0Z
 
 | | DICTYOSTELIUM MYOSIN S1DC (MOTOR DOMAIN FRAGMENT) COMPLEXED WITH P-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE. | | Descriptor: | MAGNESIUM ION, MYOSIN, P-NITROPHENYL AMINOETHYLDIPHOSPHATE BERYLLIUM TRIFLUORIDE | | Authors: | Gulick, A.M, Bauer, C.B, Thoden, J.B, Pate, E, Yount, R.G, Rayment, I. | | Deposit date: | 1999-09-15 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the Dictyostelium discoideum myosin motor domain with six non-nucleotide analogs.
J.Biol.Chem., 275, 2000
|
|
1ONJ
 
 | | Crystal structure of Atratoxin-b from Chinese cobra venom of Naja atra | | Descriptor: | Cobrotoxin b, SULFATE ION | | Authors: | Lou, X, Tu, X, Pan, G, Xu, C, Fan, R, Lu, W, Deng, W, Rao, P, Teng, M, Niu, L. | | Deposit date: | 2003-02-28 | | Release date: | 2004-02-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.555 Å) | | Cite: | Purification, N-terminal sequencing, crystallization and preliminary structural determination of atratoxin-b, a short-chain alpha-neurotoxin from Naja atra venom.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
8DRX
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp10-nsp11 (C10) cut site sequence (form 2) | | Descriptor: | Fusion protein of 3C-like proteinase nsp5 and nsp10-nsp11 (C10) cut site, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
5KVG
 
 | | Zika specific antibody, ZV-67, bound to ZIKA envelope DIII | | Descriptor: | CHLORIDE ION, ZIKA Envelope DIII, ZV-67 Antibody Fab Heavy Chain, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
3KCS
 
 | | Crystal structure of PAmCherry1 in the dark state | | Descriptor: | PAmCherry1 protein | | Authors: | Malashkevich, V.N, Subach, F.V, Zencheck, W.D, Xiao, H, Filonov, G.S, Almo, S.C, Verkhusha, V.V. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8DRT
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8BIS
 
 | | Crystal structure of cystathionine gamma-lyase from Toxoplasma gondii in complex with DL-propargylglycine | | Descriptor: | (2S)-2-aminopent-4-enoic acid, Cystathionine beta-lyase, putative, ... | | Authors: | Fernandez-Rodriguez, C, Conter, C, Oyenarte, I, Favretto, F, Quintana, I, Martinez-Chantar, M.L, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2022-11-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.266 Å) | | Cite: | Structural basis of the inhibition of cystathionine gamma-lyase from Toxoplasma gondii by propargylglycine and cysteine.
Protein Sci., 32, 2023
|
|
6MRQ
 
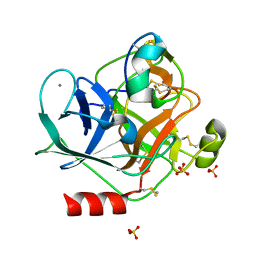 | | Structure of ToPI1 inhibitor from Tityus obscurus scorpion venom in complex with trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Fernandes, J.C, Mourao, C.B.F, Schwartz, E.F, Barbosa, J.A.R.G. | | Deposit date: | 2018-10-15 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Head-to-Tail Cyclization after Interaction with Trypsin: A Scorpion Venom Peptide that Resembles Plant Cyclotides.
J.Med.Chem., 63, 2020
|
|
1F0M
 
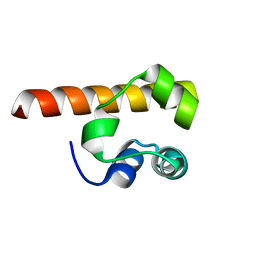 | | MONOMERIC STRUCTURE OF THE HUMAN EPHB2 SAM (STERILE ALPHA MOTIF) DOMAIN | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2 | | Authors: | Thanos, C.D, Faham, S, Goodwill, K.E, Cascio, D, Phillips, M, Bowie, J.U. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Monomeric structure of the human EphB2 sterile alpha motif domain.
J.Biol.Chem., 274, 1999
|
|
3KBL
 
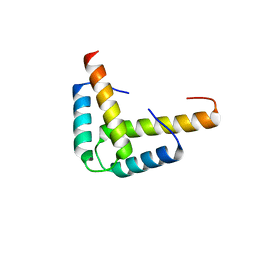 | | Crystal structure of the GLD-1 homodimerization domain from Caenorhabditis elegans N169A mutant at 2.28 A resolution | | Descriptor: | Female germline-specific tumor suppressor gld-1 | | Authors: | Beuck, C, Szymczyna, B.R, Kerkow, D.E, Carmel, A.B, Columbus, L, Stanfield, R.L, Williamson, J.R. | | Deposit date: | 2009-10-20 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Structure, 18, 2010
|
|
8BIX
 
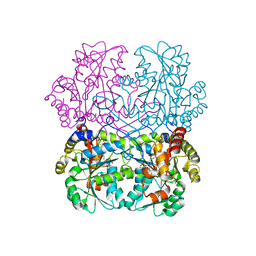 | | Cystathionine gamma-lyase N360S mutant from Toxoplasma gondii in complex with cystathionine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Cystathionine beta-lyase, putative, ... | | Authors: | Fernandez-Rodriguez, C, Conter, C, Oyenarte, I, Favretto, F, Quintana, I, Martinez-Chantar, M.L, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2022-11-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis of the inhibition of cystathionine gamma-lyase from Toxoplasma gondii by propargylglycine and cysteine.
Protein Sci., 32, 2023
|
|
6XJ6
 
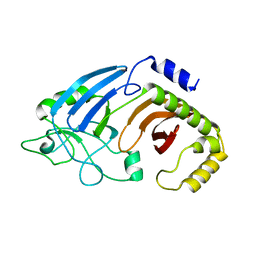 | |
2QM7
 
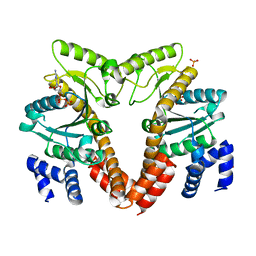 | | MeaB, A Bacterial Homolog of MMAA, Bound to GDP | | Descriptor: | GTPase/ATPase, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Hubbard, P.A, Padovani, D, Labunska, T, Mahlstedt, S.A, Banerjee, R, Drennan, C.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and mutagenesis of the metallochaperone MeaB: insight into the causes of methylmalonic aciduria.
J.Biol.Chem., 282, 2007
|
|
1PAA
 
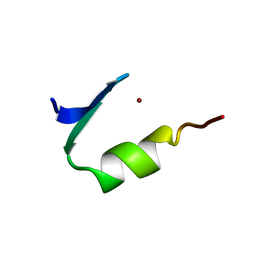 | | STRUCTURE OF A HISTIDINE-X4-HISTIDINE ZINC FINGER DOMAIN: INSIGHTS INTO ADR1-UAS1 PROTEIN-DNA RECOGNITION | | Descriptor: | YEAST TRANSCRIPTION FACTOR ADR1, ZINC ION | | Authors: | Bernstein, B.E, Hoffman, R.C, Horvath, S.J, Herriott, J.R, Klevit, R.E. | | Deposit date: | 1994-07-15 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a histidine-X4-histidine zinc finger domain: insights into ADR1-UAS1 protein-DNA recognition.
Biochemistry, 33, 1994
|
|
5DXZ
 
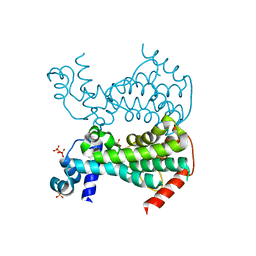 | | Crystal of AmtR from Corynebacterium glutamicum | | Descriptor: | SULFATE ION, TetR family transcriptional regulator | | Authors: | Palanca, C, Rubio, V. | | Deposit date: | 2015-09-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of AmtR, the global nitrogen regulator of Corynebacterium glutamicum, in free and DNA-bound forms.
Febs J., 283, 2016
|
|
2YUW
 
 | |
8DS0
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp14-nsp15 (C14) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS2
 
 | | Structure of SARS-CoV-2 Mpro in complex with the nsp13-nsp14 (C13) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
6ZXI
 
 | | Crystal Structure of the OXA-48 Carbapenem-Hydrolyzing Class D beta-Lactamase in Complex with the DBO inhibitor ANT3310 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CARBON DIOXIDE, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2020-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of ANT3310 , a Novel Broad-Spectrum Serine beta-Lactamase Inhibitor of the Diazabicyclooctane Class, Which Strongly Potentiates Meropenem Activity against Carbapenem-Resistant Enterobacterales and Acinetobacter baumannii.
J.Med.Chem., 63, 2020
|
|
6D42
 
 | |
2R0D
 
 | | Crystal Structure of Autoinhibited Form of Grp1 Arf GTPase Exchange Factor | | Descriptor: | Cytohesin-3, DI(HYDROXYETHYL)ETHER, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ... | | Authors: | DiNitto, J.P, Delprato, A, Gabe Lee, M.T, Cronin, T.C, Huang, S, Guilherme, A, Czech, M.P, Lambright, D.G. | | Deposit date: | 2007-08-18 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis and Mechanism of Autoregulation in 3-Phosphoinositide-Dependent Grp1 Family Arf GTPase Exchange Factors.
Mol.Cell, 28, 2007
|
|
6W4S
 
 | | Structure of apo human ferroportin in lipid nanodisc | | Descriptor: | Fab45D8 Heavy Chain, Fab45D8 Light Chain, Solute carrier family 40 member 1 | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
1JME
 
 | | Crystal Structure of Phe393His Cytochrome P450 BM3 | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ost, T.W.B, Munro, A.W, Mowat, C.G, Pesseguiero, A, Fulco, A.J, Cho, A.K, Cheesman, M.A, Walkinshaw, M.D, Chapman, S.K. | | Deposit date: | 2001-07-18 | | Release date: | 2001-11-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and spectroscopic analysis of the F393H mutant of flavocytochrome P450 BM3.
Biochemistry, 40, 2001
|
|
