6U5V
 
 | |
6U31
 
 | | The crystal structure of 4-(1H-imidazol-1-yl)benzoate-bound CYP199A4 | | Descriptor: | 4-(1H-imidazol-1-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | To Be, or Not to Be, an Inhibitor: A Comparison of Azole Interactions with and Oxidation by a Cytochrome P450 Enzyme.
Inorg.Chem., 61, 2022
|
|
3OUI
 
 | | PHD2-R717 with 40787422 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Egl nine homolog 1, ... | | Authors: | Arakaki, T.L, Kim, H. | | Deposit date: | 2010-09-14 | | Release date: | 2010-12-01 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Benzimidazole-2-pyrazole HIF Prolyl 4-Hydroxylase Inhibitors as Oral Erythropoietin Secretagogues.
ACS Med Chem Lett, 1, 2010
|
|
3OUZ
 
 | | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, D-MALATE, ... | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal Structure of Biotin Carboxylase-ADP complex from Campylobacter jejuni
TO BE PUBLISHED
|
|
2MIB
 
 | |
8G22
 
 | | Crystal Structure of the dTDP-4-dehydrorhamnose Reductase from Streptococcus pneumoniae. | | Descriptor: | dTDP-4-dehydrorhamnose reductase | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of the dTDP-4-dehydrorhamnose Reductase from Streptococcus pneumoniae.
To Be Published
|
|
3P5V
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
8DNK
 
 | | Crystal structure of human KRAS G12C covalently bound with Taiho WO2020/085493A1 compound 6 | | Descriptor: | 2-{[(5-tert-butyl-6-chloro-1H-indazol-3-yl)amino]methyl}-4-chloro-1-methyl-N-(1-propanoylazetidin-3-yl)-1H-imidazole-5-carboxamide, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2022-07-11 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Modeling receptor flexibility in the structure-based design of KRAS G12C inhibitors.
J.Comput.Aided Mol.Des., 36, 2022
|
|
8DNJ
 
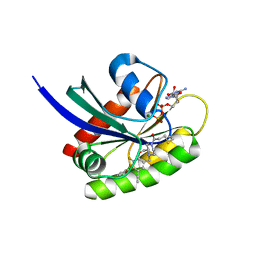 | | Crystal structure of human KRAS G12C covalently bound with AstraZeneca WO2020/178282A1 compound 76 | | Descriptor: | 1-[(5S,9P,12aR)-9-(2-chloro-6-hydroxyphenyl)-8-ethynyl-10-fluoro-3,4,12,12a-tetrahydro-6H-pyrazino[2,1-c][1,4]benzoxazepin-2(1H)-yl]propan-1-one, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Mohr, C. | | Deposit date: | 2022-07-11 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Modeling receptor flexibility in the structure-based design of KRAS G12C inhibitors.
J.Comput.Aided Mol.Des., 36, 2022
|
|
4B05
 
 | | Preclinical characterization of AZD3839, a novel clinical candidate BACE1 inhibitor for the treatment of Alzheimer Disease | | Descriptor: | (1S)-1-[2-(difluoromethyl)pyridin-4-yl]-4-fluoro-1-(3-pyrimidin-5-ylphenyl)-1H-isoindol-3-amine, ACETATE ION, BETA-SECRETASE 1, ... | | Authors: | Jeppsson, F, Eketjall, S, Janson, J, Karlstrom, S, Gustavsson, S, Olsson, L.L, Radesater, A.C, Ploeger, B, Cebers, G, Kolmodin, K, Swahn, B.M, von Berg, S, Bueters, T, Falting, J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of AZD3839, a potent and selective BACE1 inhibitor clinical candidate for the treatment of Alzheimer disease.
J. Biol. Chem., 287, 2012
|
|
6UWI
 
 | | Crystal structure of the Clostridium difficile translocase CDTb | | Descriptor: | ADP-ribosyltransferase binding component, CALCIUM ION | | Authors: | Pozharski, E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the cell-binding component of theClostridium difficilebinary toxin reveals a di-heptamer macromolecular assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3ONW
 
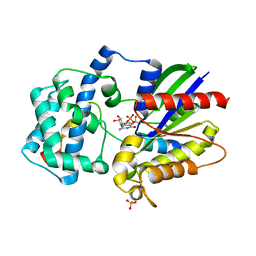 | | Structure of a G-alpha-i1 mutant with enhanced affinity for the RGS14 GoLoco motif. | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, Regulator of G-protein signaling 14, ... | | Authors: | Bosch, D, Kimple, A.J, Sammond, D.W, Miley, M.J, Machius, M, Kuhlman, B, Willard, F.S, Siderovski, D.P. | | Deposit date: | 2010-08-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Determinants of Affinity Enhancement between GoLoco Motifs and G-Protein {alpha} Subunit Mutants.
J.Biol.Chem., 286, 2011
|
|
8GJX
 
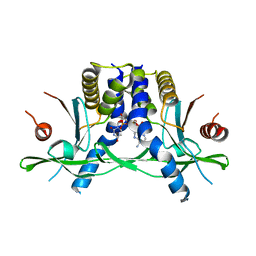 | | Structure of the human STING receptor bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Morehouse, B.R, Li, Y, Slavik, K.M, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJZ
 
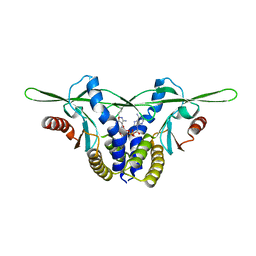 | | Structure of a STING receptor from S. pistillata Sp-STING1 bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8E80
 
 | | Structure of LRRK2-CHK1 10-pt. mutant complex with heteroaryl-1H-indazole LRRK2 inhibitor 14 | | Descriptor: | 2-[(1r,4r)-2-{(6P)-6-[(6M)-6-(1H-pyrazol-5-yl)-1H-indazol-1-yl]pyrimidin-4-yl}-2-azabicyclo[2.1.1]hexan-4-yl]propan-2-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery and Optimization of Potent, Selective, and Brain-Penetrant 1-Heteroaryl-1 H -Indazole LRRK2 Kinase Inhibitors for the Treatment of Parkinson's Disease.
J.Med.Chem., 65, 2022
|
|
8GU7
 
 | |
4ASU
 
 | | F1-ATPase in which all three catalytic sites contain bound nucleotide, with magnesium ion released in the Empty site | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE SUBUNIT ALPHA, MITOCHONDRIAL, ... | | Authors: | Rees, D.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2012-05-03 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Evidence of a New Catalytic Intermediate in the Pathway of ATP Hydrolysis by F1-ATPase from Bovine Heart Mitochondria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3Q34
 
 | |
8GO3
 
 | |
8DSN
 
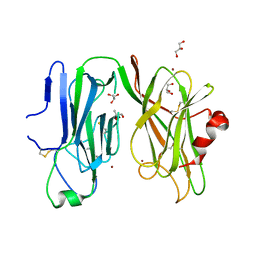 | | Peptidylglycine alpha hydroxylating monoxygenase, Q272A | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidylglycine alpha-amidating monooxygenase | | Authors: | Arias, R.J, Blackburn, N.J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New structures reveal flexible dynamics between the subdomains of peptidylglycine monooxygenase. Implications for an open to closed mechanism.
Protein Sci., 32, 2023
|
|
8DSJ
 
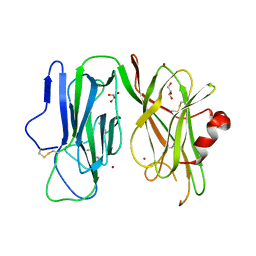 | | Peptidylglycine alpha hydroxylating monooxygenase anaerobic | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidylglycine alpha-amidating monooxygenase | | Authors: | Arias, R.J, Blackburn, N.J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | New structures reveal flexible dynamics between the subdomains of peptidylglycine monooxygenase. Implications for an open to closed mechanism.
Protein Sci., 32, 2023
|
|
8DSL
 
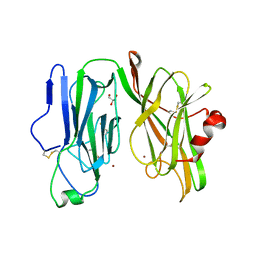 | | Peptidylglycine alpha hydroxylating monooxygenase, Q272E | | Descriptor: | COPPER (II) ION, GLYCEROL, Peptidylglycine alpha-amidating monooxygenase | | Authors: | Arias, R.J, Blackburn, N.J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | New structures reveal flexible dynamics between the subdomains of peptidylglycine monooxygenase. Implications for an open to closed mechanism.
Protein Sci., 32, 2023
|
|
6UWT
 
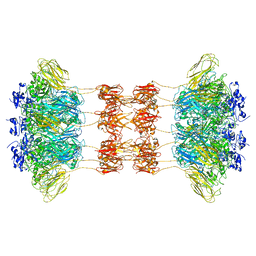 | |
2M43
 
 | | DNA containing a cluster of 8-oxo-guanine and abasic site lesion: alpha anomer (AP6, 8OG 14) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*CP*(ORP)P*CP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*(8OG)P*TP*GP*GP*GP*AP*GP*CP*G)-3') | | Authors: | Zalesak, J, Jourdan, M, Constant, J, Lourdin, M. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions.
J.Mol.Biol., 426, 2014
|
|
2LYR
 
 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
