6DAR
 
 | | Discovery of Potent 2-Aryl-6,7-Dihydro-5HPyrrolo[ 1,2-a]imidazoles as WDR5 WIN-site Inhibitors Using Fragment-Based Methods and Structure-Based Design | | Descriptor: | DIMETHYL SULFOXIDE, N-(cyclopropylmethyl)-N-{[3-(6,7-dihydro-5H-pyrrolo[1,2-a]imidazol-2-yl)phenyl]methyl}-3-methoxybenzamide, SULFATE ION, ... | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2018-05-01 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
6DAS
 
 | | Discovery of Potent 2-Aryl-6,7-Dihydro-5HPyrrolo[ 1,2-a]imidazoles as WDR5 WIN-site Inhibitors Using Fragment-Based Methods and Structure-Based Design | | Descriptor: | N-[(1R)-6-(6,7-dihydro-5H-pyrrolo[1,2-a]imidazol-2-yl)-2,3-dihydro-1H-inden-1-yl]-3-methoxy-4-methylbenzamide, WD repeat-containing protein 5 | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2018-05-01 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
6DF6
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 16ab | | Descriptor: | (8R)-8-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-1,8-dihydro-2H-[1]benzopyrano[4,3-d][1]benzoxepine-5,11-diol, Estrogen receptor, GLYCEROL | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Ortwine, D.F, Nettles, K.W, Nwachukwu, J.C. | | Deposit date: | 2018-05-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unexpected equivalent potency of a constrained chromene enantiomeric pair rationalized by co-crystal structures in complex with estrogen receptor alpha.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6DAK
 
 | | Discovery of Potent 2-Aryl-6,7-Dihydro-5HPyrrolo[ 1,2-a]imidazoles as WDR5 WIN-site Inhibitors Using Fragment-Based Methods and Structure-Based Design | | Descriptor: | DIMETHYL SULFOXIDE, N-{[3-(6,7-dihydro-5H-pyrrolo[1,2-a]imidazol-2-yl)phenyl]methyl}benzamide, WD repeat-containing protein 5 | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2018-05-01 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
5OBU
 
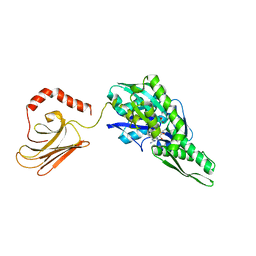 | | Mycoplasma genitalium DnaK deletion mutant lacking SBDalpha in complex with AMPPNP. | | Descriptor: | Chaperone protein DnaK, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
6DCE
 
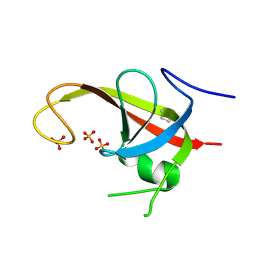 | | X-ray structure of FIP200 claw domain | | Descriptor: | RB1-inducible coiled-coil protein 1, SULFATE ION | | Authors: | Su, M.-Y, Hurley, J.H. | | Deposit date: | 2018-05-05 | | Release date: | 2019-03-06 | | Last modified: | 2019-05-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates.
Mol. Cell, 74, 2019
|
|
6DAI
 
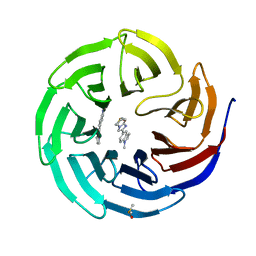 | | Discovery of Potent 2-Aryl-6,7-Dihydro-5HPyrrolo[ 1,2-a]imidazoles as WDR5 WIN-site Inhibitors Using Fragment-Based Methods and Structure-Based Design | | Descriptor: | 6-(6,7-dihydro-5H-pyrrolo[1,2-a]imidazol-2-yl)-1-methylindoline, DIMETHYL SULFOXIDE, WD repeat-containing protein 5 | | Authors: | Phan, J, Fesik, S.W. | | Deposit date: | 2018-05-01 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of Potent 2-Aryl-6,7-dihydro-5 H-pyrrolo[1,2- a]imidazoles as WDR5-WIN-Site Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
6CBU
 
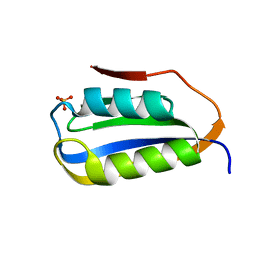 | | Crystal structure of C4S3: A computationally designed immunogen to target Carbohydrate-Occluded Epitopes on the HIV envelope | | Descriptor: | Acylphosphatase-1, SULFATE ION | | Authors: | Zhu, C, Ke, H.M, Swanstrom, R, Dokholyan, N.V. | | Deposit date: | 2018-02-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rationally designed carbohydrate-occluded epitopes elicit HIV-1 Env-specific antibodies.
Nat Commun, 10, 2019
|
|
6DEJ
 
 | |
6DFN
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 16aa | | Descriptor: | (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, (8S)-8-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-1,8-dihydro-2H-[1]benzopyrano[4,3-d][1]benzoxepine-5,11-diol, Estrogen receptor, ... | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Ortwine, D.F, Nettles, K.W, Nwachukwu, J.C. | | Deposit date: | 2018-05-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected equivalent potency of a constrained chromene enantiomeric pair rationalized by co-crystal structures in complex with estrogen receptor alpha.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6E1A
 
 | | Menin bound to M-89 | | Descriptor: | (1S,2R)-2-[(4S)-2-methyl-4-{1-[(1-{4-[(pyridin-4-yl)sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}-1,2,3,4-tetrahydroisoquinolin-4-yl]cyclopentyl methylcarbamate, Menin, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2018-07-09 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery of M-89 as a Highly Potent Inhibitor of the Menin-Mixed Lineage Leukemia (Menin-MLL) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
7E3V
 
 | | Metallo beta-lactamase fold protein (cAMP free) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Lee, K.-Y, Kim, D.-G, Lee, B.-J. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional studies of SAV1707 from Staphylococcus aureus elucidate its distinct metal-dependent activity and a crucial residue for catalysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7CBJ
 
 | | Crystal structure of PDE4D catalytic domain in complex with compound 36 | | Descriptor: | (1S)-1-[(7-chloranyl-1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydro-1H-isoquinoline-2-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, and biological evaluation of tetrahydroisoquinolines derivatives as novel, selective PDE4 inhibitors for antipsoriasis treatment.
Eur.J.Med.Chem., 211, 2021
|
|
4AWI
 
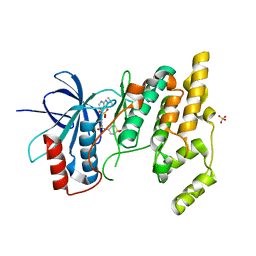 | | Human Jnk1alpha kinase with 4-phenyl-7-azaindole IKK2 inhibitor. | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 8, N-(1,1-dioxidotetrahydro-2H-thiopyran-4-yl)-4-[2-(1-methylethyl)-1H-pyrrolo[2,3-b]pyridin-4-yl]benzenesulfonamide, SULFATE ION | | Authors: | Chung, C, Vicentini, G, Liddle, J, Bamborough, P. | | Deposit date: | 2012-06-03 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | 4-Phenyl-7-Azaindoles as Potent, Selective and Bioavailable Ikk2 Inhibitors Demonstrating Good in Vivo Efficacy.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4BWO
 
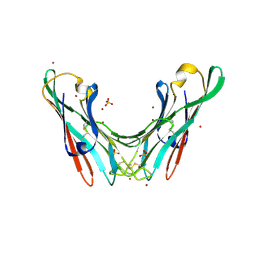 | | The FedF adhesin from entrrotoxigenic Escherichia coli is a sulfate- binding lectin | | Descriptor: | BROMIDE ION, F18 FIMBRIAL ADHESIN AC, SULFATE ION | | Authors: | Lonardi, E, Moonens, K, Buts, L, de Boer, A.R, Olsson, J.D.M, Weiss, M.S, Fabre, E, Guerardel, Y, Deelder, A.M, Oscarson, S, Wuhrer, M, Bouckaert, J. | | Deposit date: | 2013-07-03 | | Release date: | 2013-08-28 | | Last modified: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia Coli
Biology, 2, 2013
|
|
4BL9
 
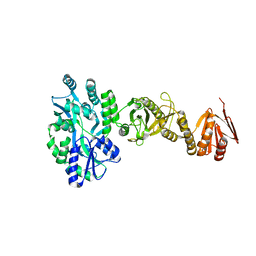 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form I) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BLB
 
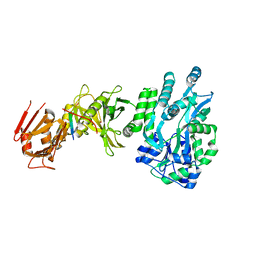 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI1p complex | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, ZINC FINGER PROTEIN GLI1, ... | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7F2L
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 18a | | Descriptor: | (~{E})-4-[9-[(4-fluorophenyl)methoxy]-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-6-oxidanylidene-pyrano[3,2-b]xanthen-5-yl]oxybut-2-enoic acid, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2021-06-11 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.10111427 Å) | | Cite: | Mangostanin Derivatives as Novel and Orally Active Phosphodiesterase 4 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis with Improved Safety.
J.Med.Chem., 64, 2021
|
|
7F2M
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 18d | | Descriptor: | (~{Z})-4-[9-[(4-fluorophenyl)methoxy]-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-6-oxidanylidene-pyrano[3,2-b]xanthen-5-yl]oxybut-2-enoic acid, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2021-06-11 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20004153 Å) | | Cite: | Mangostanin Derivatives as Novel and Orally Active Phosphodiesterase 4 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis with Improved Safety.
J.Med.Chem., 64, 2021
|
|
7F2K
 
 | | Crystal structure of PDE4D catalytic domain complexed with compound 17a | | Descriptor: | (~{E})-4-[8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-9-oxidanyl-6-oxidanylidene-pyrano[3,2-b]xanthen-5-yl]oxybut-2-enoic acid, Isoform 3 of cAMP-specific 3',5'-cyclic phosphodiesterase 4D, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2021-06-11 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.10001969 Å) | | Cite: | Mangostanin Derivatives as Novel and Orally Active Phosphodiesterase 4 Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis with Improved Safety.
J.Med.Chem., 64, 2021
|
|
4BLA
 
 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form II) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5OY4
 
 | | GSK3beta complex with N-(6-(3,4-dihydroxyphenyl)-1H-pyrazolo[3,4-b]pyridin-3-yl)acetamide | | Descriptor: | Glycogen synthase kinase-3 beta, Proto-oncogene FRAT1, SULFATE ION, ... | | Authors: | Bax, B.D, Convery, M.A. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | From PIM1 to PI3K delta via GSK3 beta : Target Hopping through the Kinome.
ACS Med Chem Lett, 8, 2017
|
|
7FIC
 
 | |
4AW8
 
 | | X-ray structure of ZinT from Salmonella enterica in complex with zinc ion and PEG | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Metal-binding protein ZinT, SODIUM ION, ... | | Authors: | Alaleona, F, Ilari, A, Battistoni, A, Petrarca, P, Chiancone, E. | | Deposit date: | 2012-06-01 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Salmonella Enterica Zint Structure, Zinc Affinity and Interaction with the High-Affinity Uptake Protein Znua Provide Insight Into the Management of Periplasmic Zinc.
Biochim.Biophys.Acta, 1840, 2014
|
|
4AYH
 
 | | The X-ray structure of zinc bound ZinT | | Descriptor: | METAL-BINDING PROTEIN YODA, SULFATE ION, ZINC ION | | Authors: | Alaleona, F, Ilari, A, Battistoni, A, Petrarca, P, Chiancone, E. | | Deposit date: | 2012-06-21 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Salmonella Enterica Zint Structure, Zinc Affinity and Interaction with the High-Affinity Uptake Protein Znua Provide Insight Into the Management of Periplasmic Zinc.
Biochim.Biophys.Acta, 1840, 2014
|
|
